Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs
- PMID: 26814032
- PMCID: PMC4737851
- DOI: 10.1038/ncomms10476
Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs
Abstract
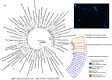
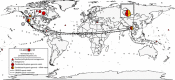
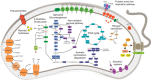
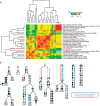
Analysis of the increasing wealth of metagenomic data collected from diverse environments can lead to the discovery of novel branches on the tree of life. Here we analyse 5.2 Tb of metagenomic data collected globally to discover a novel bacterial phylum ('Candidatus Kryptonia') found exclusively in high-temperature pH-neutral geothermal springs. This lineage had remained hidden as a taxonomic 'blind spot' because of mismatches in the primers commonly used for ribosomal gene surveys. Genome reconstruction from metagenomic data combined with single-cell genomics results in several high-quality genomes representing four genera from the new phylum. Metabolic reconstruction indicates a heterotrophic lifestyle with conspicuous nutritional deficiencies, suggesting the need for metabolic complementarity with other microbes. Co-occurrence patterns identifies a number of putative partners, including an uncultured Armatimonadetes lineage. The discovery of Kryptonia within previously studied geothermal springs underscores the importance of globally sampled metagenomic data in detection of microbial novelty, and highlights the extraordinary diversity of microbial life still awaiting discovery.
Figures

References
-
- Yarza P. et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645 (2014). - PubMed
-
- Brown C. T. et al. Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523, 208–211 (2015). - PubMed
-
- Iverson V. et al. Untangling genomes from metagenomes: revealing an uncultured class of marine euryarchaeota. Science 335, 587–590 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

