The Tumor-suppressive Small GTPase DiRas1 Binds the Noncanonical Guanine Nucleotide Exchange Factor SmgGDS and Antagonizes SmgGDS Interactions with Oncogenic Small GTPases
- PMID: 26814130
- PMCID: PMC4813585
- DOI: 10.1074/jbc.M115.696831
The Tumor-suppressive Small GTPase DiRas1 Binds the Noncanonical Guanine Nucleotide Exchange Factor SmgGDS and Antagonizes SmgGDS Interactions with Oncogenic Small GTPases
Erratum in
-
The tumor-suppressive small GTPase DiRas1 binds the noncanonical guanine nucleotide exchange factor SmgGDS and antagonizes SmgGDS interactions with oncogenic small GTPases.J Biol Chem. 2016 May 13;291(20):10948. doi: 10.1074/jbc.A115.696831. J Biol Chem. 2016. PMID: 27197236 Free PMC article. No abstract available.
Abstract
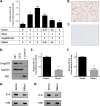
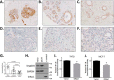
The small GTPase DiRas1 has tumor-suppressive activities, unlike the oncogenic properties more common to small GTPases such as K-Ras and RhoA. Although DiRas1 has been found to be a tumor suppressor in gliomas and esophageal squamous cell carcinomas, the mechanisms by which it inhibits malignant phenotypes have not been fully determined. In this study, we demonstrate that DiRas1 binds to SmgGDS, a protein that promotes the activation of several oncogenic GTPases. In silico docking studies predict that DiRas1 binds to SmgGDS in a manner similar to other small GTPases. SmgGDS is a guanine nucleotide exchange factor for RhoA, but we report here that SmgGDS does not mediate GDP/GTP exchange on DiRas1. Intriguingly, DiRas1 acts similarly to a dominant-negative small GTPase, binding to SmgGDS and inhibiting SmgGDS binding to other small GTPases, including K-Ras4B, RhoA, and Rap1A. DiRas1 is expressed in normal breast tissue, but its expression is decreased in most breast cancers, similar to its family member DiRas3 (ARHI). DiRas1 inhibits RhoA- and SmgGDS-mediated NF-κB transcriptional activity in HEK293T cells. We also report that DiRas1 suppresses basal NF-κB activation in breast cancer and glioblastoma cell lines. Taken together, our data support a model in which DiRas1 expression inhibits malignant features of cancers in part by nonproductively binding to SmgGDS and inhibiting the binding of other small GTPases to SmgGDS.
Keywords: Di-Ras1; GTPase Kras (KRAS); NF-kappaB (NF-KB); Rap1GDS1; Ras homolog gene family, member A (RhoA); Ras-related protein 1 (Rap1); Rig; SmgGDS; breast cancer; glioblastoma.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Bos J. L. (1989) ras oncogenes in human cancer: a review. Cancer Res. 49, 4682–4689 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

