Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data--The Influence of Different Parameters in a Routine Clinical Microbiology Laboratory
- PMID: 26814675
- PMCID: PMC4729434
- DOI: 10.1371/journal.pone.0147965
Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data--The Influence of Different Parameters in a Routine Clinical Microbiology Laboratory
Abstract
Introduction: Many clinical microbiology laboratories report on cumulative antimicrobial susceptibility testing (cAST) data on a regular basis. Criteria for generation of cAST reports, however, are often obscure and inconsistent. Whereas the CLSI has published a guideline for analysis and presentation of cAST data, national guidelines directed at clinical microbiology laboratories are not available in Europe. Thus, we sought to describe the influence of different parameters in the process of cAST data analysis in the setting of a German routine clinical microbiology laboratory during 2 consecutive years.
Material and methods: We developed various program scripts to assess the consequences ensuing from different algorithms for calculation of cumulative antibiograms from the data collected in our clinical microbiology laboratory in 2013 and 2014.
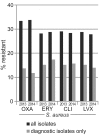
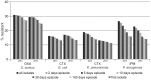
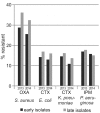
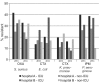
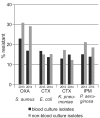
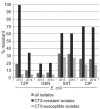
Results: One of the most pronounced effects was caused by exclusion of screening cultures for multi-drug resistant organisms which decreased the MRSA rate in some cases to one third. Dependent on the handling of duplicate isolates, i.e. isolates of the same species recovered from successive cultures on the same patient during the time period analyzed, we recorded differences in resistance rates of up to 5 percentage points for S. aureus, E. coli and K. pneumoniae and up to 10 percentage points for P. aeruginosa. Stratification by site of care and specimen type, testing of antimicrobials selectively on resistant isolates, change of interpretation rules and analysis at genus level instead of species level resulted in further changes of calculated antimicrobial resistance rates.
Conclusion: The choice of parameters for cAST data analysis may have a substantial influence on calculated antimicrobial resistance rates. Consequently, comparability of cAST reports from different clinical microbiology laboratories may be limited. We suggest that laboratories communicate the strategy used for cAST data analysis as long as national guidelines for standardized cAST data analysis and reporting do not exist in Europe.
Conflict of interest statement
Figures
Similar articles
-
Comparison of Clinical Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing guidelines for the interpretation of antibiotic susceptibility at a University teaching hospital in Nairobi, Kenya: a cross-sectional study.Ann Clin Microbiol Antimicrob. 2016 Apr 11;15:21. doi: 10.1186/s12941-016-0135-3. Ann Clin Microbiol Antimicrob. 2016. PMID: 27068515 Free PMC article.
-
Temporal changes in bacterial resistance in German intensive care units, 2001-2003: data from the SARI (surveillance of antimicrobial use and antimicrobial resistance in intensive care units) project.J Hosp Infect. 2005 Aug;60(4):348-52. doi: 10.1016/j.jhin.2004.12.027. J Hosp Infect. 2005. PMID: 15923060
-
Influence of analysis conditions for antimicrobial susceptibility test data on susceptibility rates.PLoS One. 2020 Jun 23;15(6):e0235059. doi: 10.1371/journal.pone.0235059. eCollection 2020. PLoS One. 2020. PMID: 32574199 Free PMC article.
-
[Trends of causative and antimicrobial resistant organisms among patients with hematological malignancies in Japan].Rinsho Ketsueki. 2011 Apr;52(4):177-81. Rinsho Ketsueki. 2011. PMID: 21566402 Review. Japanese. No abstract available.
-
Systematic review of antibiograms: A National Laboratory System approach for improving antimicrobial susceptibility testing practices in Michigan.Public Health Rep. 2010 May-Jun;125 Suppl 2(Suppl 2):63-72. doi: 10.1177/00333549101250S208. Public Health Rep. 2010. PMID: 20518446 Free PMC article.
Cited by
-
Microbiology Investigation Criteria for Reporting Objectively (MICRO): a framework for the reporting and interpretation of clinical microbiology data.BMC Med. 2019 Mar 29;17(1):70. doi: 10.1186/s12916-019-1301-1. BMC Med. 2019. PMID: 30922309 Free PMC article.
-
Microbiological Profiles and Resistance Patterns in Pediatrics With Cancer: An 8-Year Study at a Comprehensive Cancer Center in Jordan.Cancer Rep (Hoboken). 2025 Mar;8(3):e70132. doi: 10.1002/cnr2.70132. Cancer Rep (Hoboken). 2025. PMID: 40065500 Free PMC article.
-
Determinants of methicillin-resistant Staphylococcus aureus (MRSA) prevalence in the Asia-Pacific region: A systematic review and meta-analysis.J Glob Antimicrob Resist. 2019 Mar;16:17-27. doi: 10.1016/j.jgar.2018.08.014. Epub 2018 Aug 23. J Glob Antimicrob Resist. 2019. PMID: 30145271 Free PMC article.
-
The antibiogram: key considerations for its development and utilization.JAC Antimicrob Resist. 2021 May 25;3(2):dlab060. doi: 10.1093/jacamr/dlab060. eCollection 2021 Jun. JAC Antimicrob Resist. 2021. PMID: 34223122 Free PMC article. Review.
-
Diagnostic Accuracy of Hospital Antibiograms in Predicting the Risk of Antimicrobial Resistance in Enterobacteriaceae Isolates: A Nationwide Multicenter Evaluation at the Veterans Health Administration.Clin Infect Dis. 2023 Nov 30;77(11):1492-1500. doi: 10.1093/cid/ciad467. Clin Infect Dis. 2023. PMID: 37658908 Free PMC article.
References
-
- CLSI. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data; Approved Guideline—Third Edition. CLSI document M39-A4. Wayne, PA: Clinical and Laboratory Standards Institute; 2009.
-
- Xu R, Polk RE, Stencel L, Lowe DK, Guharoy R, Duggal RW, et al. Antibiogram compliance in University HealthSystem Consortium participating hospitals with Clinical and Laboratory Standards Institute guidelines. Am J Health Syst Pharm. 2012;69(7):598–606. Epub 2012/03/24. 10.2146/ajhp110332 69/7/598 [pii]. . - DOI - PubMed
-
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0, 2015. http://www.eucast.org. 2015.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

