Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells
- PMID: 26814966
- PMCID: PMC4871117
- DOI: 10.1038/nature16505
Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells
Abstract
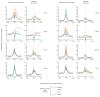
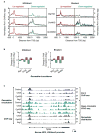
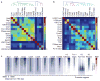
ATP-dependent chromatin remodellers allow access to DNA for transcription factors and the general transcription machinery, but whether mammalian chromatin remodellers target specific nucleosomes to regulate transcription is unclear. Here we present genome-wide remodeller-nucleosome interaction profiles for the chromatin remodellers Chd1, Chd2, Chd4, Chd6, Chd8, Chd9, Brg1 and Ep400 in mouse embryonic stem (ES) cells. These remodellers bind one or both full nucleosomes that flank micrococcal nuclease (MNase)-defined nucleosome-free promoter regions (NFRs), where they separate divergent transcription. Surprisingly, large CpG-rich NFRs that extend downstream of annotated transcriptional start sites are nevertheless bound by non-nucleosomal or subnucleosomal histone variants (H3.3 and H2A.Z) and marked by H3K4me3 and H3K27ac modifications. RNA polymerase II therefore navigates hundreds of base pairs of altered chromatin in the sense direction before encountering an MNase-resistant nucleosome at the 3' end of the NFR. Transcriptome analysis after remodeller depletion reveals reciprocal mechanisms of transcriptional regulation by remodellers. Whereas at active genes individual remodellers have either positive or negative roles via altering nucleosome stability, at polycomb-enriched bivalent genes the same remodellers act in an opposite manner. These findings indicate that remodellers target specific nucleosomes at the edge of NFRs, where they regulate ES cell transcriptional programs.
Conflict of interest statement
The authors declare no competing financial interests.
Figures









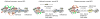
Comment in
-
Epigenomics: Custom remodelling.Nat Rev Genet. 2016 Mar;17(3):126-7. doi: 10.1038/nrg.2016.16. Epub 2016 Feb 8. Nat Rev Genet. 2016. PMID: 26852808 No abstract available.
Similar articles
-
Histone variant H2A.Z regulates nucleosome unwrapping and CTCF binding in mouse ES cells.Nucleic Acids Res. 2020 Jun 19;48(11):5939-5952. doi: 10.1093/nar/gkaa360. Nucleic Acids Res. 2020. PMID: 32392318 Free PMC article.
-
ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference.Nature. 2017 Aug 31;548(7669):607-611. doi: 10.1038/nature23671. Epub 2017 Aug 2. Nature. 2017. PMID: 28767641 Free PMC article.
-
Regulation of Nucleosome Architecture and Factor Binding Revealed by Nuclease Footprinting of the ESC Genome.Cell Rep. 2015 Oct 6;13(1):61-69. doi: 10.1016/j.celrep.2015.08.071. Epub 2015 Sep 24. Cell Rep. 2015. PMID: 26411677 Free PMC article.
-
Histone exchange, chromatin structure and the regulation of transcription.Nat Rev Mol Cell Biol. 2015 Mar;16(3):178-89. doi: 10.1038/nrm3941. Epub 2015 Feb 4. Nat Rev Mol Cell Biol. 2015. PMID: 25650798 Review.
-
Energy-driven genome regulation by ATP-dependent chromatin remodellers.Nat Rev Mol Cell Biol. 2024 Apr;25(4):309-332. doi: 10.1038/s41580-023-00683-y. Epub 2023 Dec 11. Nat Rev Mol Cell Biol. 2024. PMID: 38081975 Free PMC article. Review.
Cited by
-
ADNP Controls Gene Expression Through Local Chromatin Architecture by Association With BRG1 and CHD4.Front Cell Dev Biol. 2020 Jul 1;8:553. doi: 10.3389/fcell.2020.00553. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32714933 Free PMC article.
-
The chromatin remodeler CHD6 promotes colorectal cancer development by regulating TMEM65-mediated mitochondrial dynamics via EGF and Wnt signaling.Cell Discov. 2022 Dec 6;8(1):130. doi: 10.1038/s41421-022-00478-z. Cell Discov. 2022. PMID: 36473865 Free PMC article.
-
A molecular roadmap for the emergence of early-embryonic-like cells in culture.Nat Genet. 2018 Jan;50(1):106-119. doi: 10.1038/s41588-017-0016-5. Epub 2017 Dec 18. Nat Genet. 2018. PMID: 29255263 Free PMC article.
-
Genome-wide ATAC-see screening identifies TFDP1 as a modulator of global chromatin accessibility.Nat Genet. 2024 Mar;56(3):473-482. doi: 10.1038/s41588-024-01658-1. Epub 2024 Feb 15. Nat Genet. 2024. PMID: 38361031
-
Epigenome Regulation by Dynamic Nucleosome Unwrapping.Trends Biochem Sci. 2020 Jan;45(1):13-26. doi: 10.1016/j.tibs.2019.09.003. Epub 2019 Oct 17. Trends Biochem Sci. 2020. PMID: 31630896 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

