Critical Nucleus Structure and Aggregation Mechanism of the C-terminal Fragment of Copper-Zinc Superoxide Dismutase Protein
- PMID: 26815332
- PMCID: PMC7842942
- DOI: 10.1021/acschemneuro.5b00242
Critical Nucleus Structure and Aggregation Mechanism of the C-terminal Fragment of Copper-Zinc Superoxide Dismutase Protein
Abstract
The aggregation of the copper-zinc superoxide dismutase (SOD1) protein is linked to familial amyotrophic lateral sclerosis, a progressive neurodegenerative disease. A recent experimental study has shown that the (147)GVIGIAQ(153) SOD1 C-terminal segment not only forms amyloid fibrils in isolation but also accelerates the aggregation of full-length SOD1, while substitution of isoleucine at site 149 by proline blocks its fibril formation. Amyloid formation is a nucleation-polymerization process. In this study, we investigated the oligomerization and the nucleus structure of this heptapeptide. By performing extensive replica-exchange molecular dynamics (REMD) simulations and conventional MD simulations, we found that the GVIGIAQ hexamers can adopt highly ordered bilayer β-sheets and β-barrels. In contrast, substitution of I149 by proline significantly reduces the β-sheet probability and results in the disappearance of bilayer β-sheet structures and the increase of disordered hexamers. We identified mixed parallel-antiparallel bilayer β-sheets in both REMD and conventional MD simulations and provided the conformational transition from the experimentally observed parallel bilayer sheets to the mixed parallel-antiparallel bilayer β-sheets. Our simulations suggest that the critical nucleus consists of six peptide chains and two additional peptide chains strongly stabilize this critical nucleus. The stabilized octamer is able to recruit additional random peptides into the β-sheet. Therefore, our simulations provide insights into the critical nucleus formation and the smallest stable nucleus of the (147)GVIGIAQ(153) peptide.
Keywords: Replica exchange method; bilayer β-sheet; free energy landscape; molecular dynamics simulations; nucleus; oligomers.
Figures
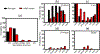









Similar articles
-
Mechanistic insight into E22Q-mutation-induced antiparallel-to-parallel β-sheet transition of Aβ16-22 fibrils: an all-atom simulation study.Phys Chem Chem Phys. 2019 Jul 17;21(28):15686-15694. doi: 10.1039/c9cp02561h. Phys Chem Chem Phys. 2019. PMID: 31271401
-
Aβ(16-22) peptides can assemble into ordered β-barrels and bilayer β-sheets, while substitution of phenylalanine 19 by tryptophan increases the population of disordered aggregates.J Phys Chem B. 2013 Sep 5;117(35):10149-60. doi: 10.1021/jp405869a. Epub 2013 Aug 26. J Phys Chem B. 2013. PMID: 23926957
-
Aβ monomers transiently sample oligomer and fibril-like configurations: ensemble characterization using a combined MD/NMR approach.J Mol Biol. 2013 Sep 23;425(18):3338-59. doi: 10.1016/j.jmb.2013.06.021. Epub 2013 Jun 25. J Mol Biol. 2013. PMID: 23811057 Free PMC article.
-
Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review.
-
Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases.Acc Chem Res. 2018 Mar 20;51(3):706-718. doi: 10.1021/acs.accounts.7b00554. Epub 2018 Mar 6. Acc Chem Res. 2018. PMID: 29508987 Free PMC article. Review.
Cited by
-
β-barrel Oligomers as Common Intermediates of Peptides Self-Assembling into Cross-β Aggregates.Sci Rep. 2018 Jul 9;8(1):10353. doi: 10.1038/s41598-018-28649-7. Sci Rep. 2018. PMID: 29985420 Free PMC article.
-
Thermo- and pH-responsive fibrillization of squid suckerin A1H1 peptide.Nanoscale. 2020 Mar 21;12(11):6307-6317. doi: 10.1039/c9nr09271d. Epub 2020 Feb 28. Nanoscale. 2020. PMID: 32108838 Free PMC article.
-
The Early Phase of β2m Aggregation: An Integrative Computational Study Framed on the D76N Mutant and the ΔN6 Variant.Biomolecules. 2019 Aug 14;9(8):366. doi: 10.3390/biom9080366. Biomolecules. 2019. PMID: 31416179 Free PMC article.
-
Evaluation of variability in high-resolution protein structures by global distance scoring.Heliyon. 2018 Feb 1;4(1):e00510. doi: 10.1016/j.heliyon.2018.e00510. eCollection 2018 Jan. Heliyon. 2018. PMID: 29560428 Free PMC article.
-
Direct Observation of β-Barrel Intermediates in the Self-Assembly of Toxic SOD128-38 and Absence in Nontoxic Glycine Mutants.J Chem Inf Model. 2021 Feb 22;61(2):966-975. doi: 10.1021/acs.jcim.0c01319. Epub 2021 Jan 14. J Chem Inf Model. 2021. PMID: 33445870 Free PMC article.
References
-
- Rowland LP, and Shneider NA (2001) Amyotrophic lateral sclerosis. N. Engl. J. Med. 344, 1688–1700. - PubMed
-
- Pasinelli P, and Brown RH (2006) Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 7, 710–723. - PubMed
-
- Cudkowicz ME, McKenna-Yasek D, Sapp PE, Chin W, Geller B, Hayden DL, Schoenfeld DA, Hosler BA, Horvitz HR, and Brown RH (1997) Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann. Neurol. 41, 210–221. - PubMed
-
- Cleveland DW, and Rothstein JD (2001) From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2, 806–819. - PubMed
-
- Fridovich I. (1975) Superoxide dismutases. Annu. Rev. Biochem. 44, 147–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

