The binding specificity of Translocated in LipoSarcoma/FUsed in Sarcoma with lncRNA transcribed from the promoter region of cyclin D1
- PMID: 26816614
- PMCID: PMC4727290
- DOI: 10.1186/s13578-016-0068-8
The binding specificity of Translocated in LipoSarcoma/FUsed in Sarcoma with lncRNA transcribed from the promoter region of cyclin D1
Abstract
Background: Translocated in LipoSarcoma (TLS, also known as FUsed in Sarcoma) is an RNA/DNA binding protein whose mutation cause amyotrophic lateral sclerosis. In previous study, we demonstrated that TLS binds to long noncoding RNA, promoter-associated ncRNA-D (pncRNA-D), transcribed from the 5' upstream region of cyclin D1 (CCND1), and inhibits the expression of CCND1.
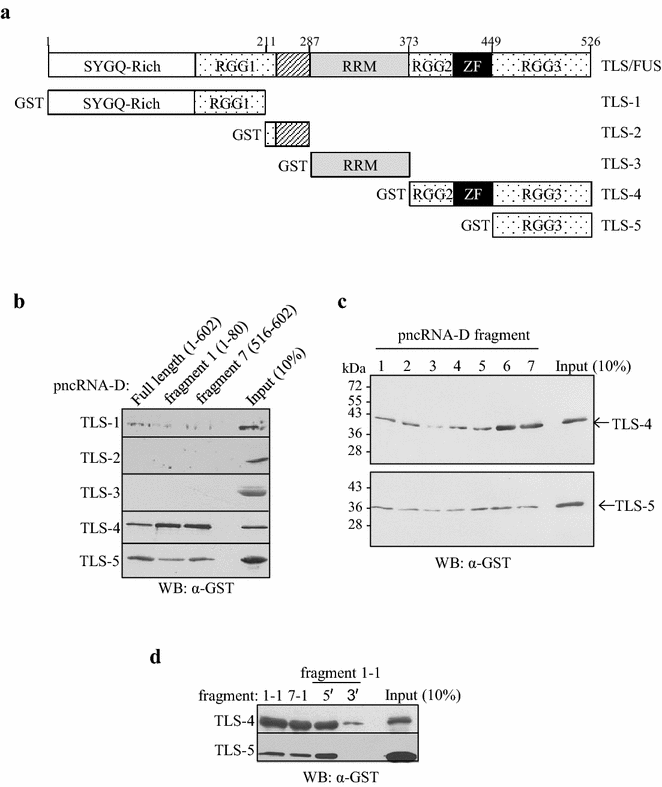
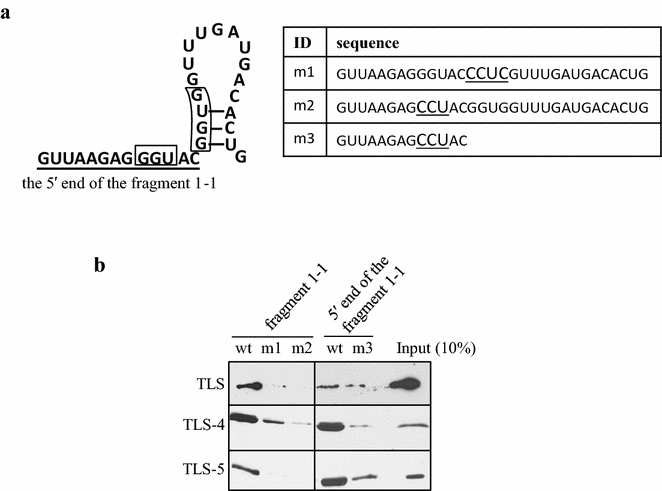
Results: In order to elucidate the binding specificity between TLS and pncRNA-D, we divided pncRNA-D into seven fragments and examined the binding with full-length TLS, TLS-RGG2-zinc finger-RGG3, and TLS-RGG3 by RNA pull down assay. As a result, TLS was able to bind to all the seven fragments, but the fragments containing reported recognition motifs (GGUG and GGU) tend to bind more solidly. The full-length TLS and TLS-RGG2-zinc finger-RGG3 showed a similar interaction with pncRNA-D, but the binding specificity of TLS-RGG3 was lower compared to the full-length TLS and TLS-RGG2-zinc finger-RGG3. Mutation in GGUG and GGU motifs dramatically decreased the binding, and unexpectedly, we could only detect weak interaction with the RNA sequence with stem loop structure.
Conclusion: The binding of TLS and pncRNA-D was affected by the presence of GGUG and GGU sequences, and the C terminal domains of TLS function in the interaction with pncRNA-D.
Keywords: Long noncoding RNA; TLS/FUS; pncRNA.
Figures



References
-
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo J-M, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. - DOI - PMC - PubMed
-
- Kwiatkowski TJ, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

