Identification of miRNA-mRNA regulatory modules by exploring collective group relationships
- PMID: 26817421
- PMCID: PMC4895272
- DOI: 10.1186/s12864-015-2300-z
Identification of miRNA-mRNA regulatory modules by exploring collective group relationships
Abstract
Background: microRNAs (miRNAs) play an essential role in the post-transcriptional gene regulation in plants and animals. They regulate a wide range of biological processes by targeting messenger RNAs (mRNAs). Evidence suggests that miRNAs and mRNAs interact collectively in gene regulatory networks. The collective relationships between groups of miRNAs and groups of mRNAs may be more readily interpreted than those between individual miRNAs and mRNAs, and thus are useful for gaining insight into gene regulation and cell functions. Several computational approaches have been developed to discover miRNA-mRNA regulatory modules (MMRMs) with a common aim to elucidate miRNA-mRNA regulatory relationships. However, most existing methods do not consider the collective relationships between a group of miRNAs and the group of targeted mRNAs in the process of discovering MMRMs. Our aim is to develop a framework to discover MMRMs and reveal miRNA-mRNA regulatory relationships from the heterogeneous expression data based on the collective relationships.
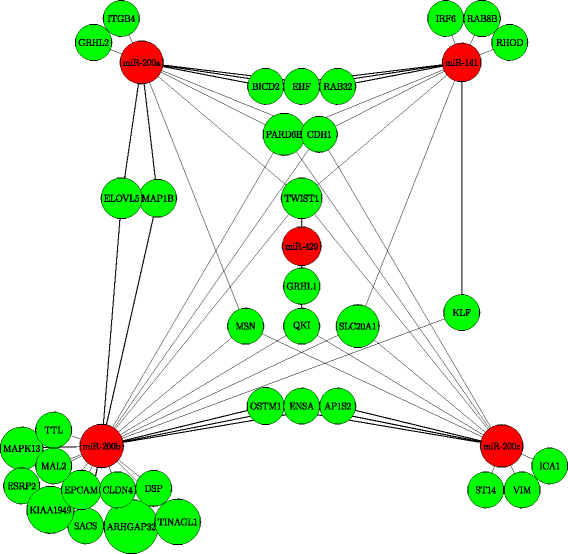
Results: We propose DIscovering COllective group RElationships (DICORE), an effective computational framework for revealing miRNA-mRNA regulatory relationships. We utilize the notation of collective group relationships to build the computational framework. The method computes the collaboration scores of the miRNAs and mRNAs on the basis of their interactions with mRNAs and miRNAs, respectively. Then it determines the groups of miRNAs and groups of mRNAs separately based on their respective collaboration scores. Next, it calculates the strength of the collective relationship between each pair of miRNA group and mRNA group using canonical correlation analysis, and the group pairs with significant canonical correlations are considered as the MMRMs. We applied this method to three gene expression datasets, and validated the computational discoveries.
Conclusions: Analysis of the results demonstrates that a large portion of the regulatory relationships discovered by DICORE is consistent with the experimentally confirmed databases. Furthermore, it is observed that the top mRNAs that are regulated by the miRNAs in the identified MMRMs are highly relevant to the biological conditions of the given datasets. It is also shown that the MMRMs identified by DICORE are more biologically significant and functionally enriched.
Figures


Similar articles
-
Identifying direct miRNA-mRNA causal regulatory relationships in heterogeneous data.J Biomed Inform. 2014 Dec;52:438-47. doi: 10.1016/j.jbi.2014.08.005. Epub 2014 Aug 30. J Biomed Inform. 2014. PMID: 25181465
-
Inferring microRNA-mRNA causal regulatory relationships from expression data.Bioinformatics. 2013 Mar 15;29(6):765-71. doi: 10.1093/bioinformatics/btt048. Epub 2013 Jan 30. Bioinformatics. 2013. PMID: 23365408
-
MIMPFC: Identifying miRNA-mRNA regulatory modules by combining phase-only correlation and improved rough-fuzzy clustering.J Bioinform Comput Biol. 2018 Feb;16(1):1750028. doi: 10.1142/S0219720017500287. Epub 2017 Dec 28. J Bioinform Comput Biol. 2018. PMID: 29281954
-
Targeting of mRNAs by multiple miRNAs: the next step.Oncogene. 2010 Apr 15;29(15):2161-4. doi: 10.1038/onc.2010.59. Epub 2010 Mar 1. Oncogene. 2010. PMID: 20190803 Review.
-
Integrative computational mRNA-miRNA interaction analyses of the autoimmune-deregulated miRNAs and well-known Th17 differentiation regulators: An attempt to discover new potential miRNAs involved in Th17 differentiation.Gene. 2015 Nov 10;572(2):153-62. doi: 10.1016/j.gene.2015.08.043. Epub 2015 Aug 22. Gene. 2015. PMID: 26307197 Review.
Cited by
-
miRModuleNet: Detecting miRNA-mRNA Regulatory Modules.Front Genet. 2022 Apr 12;13:767455. doi: 10.3389/fgene.2022.767455. eCollection 2022. Front Genet. 2022. PMID: 35495139 Free PMC article.
-
Modeling miRNA-mRNA interactions that cause phenotypic abnormality in breast cancer patients.PLoS One. 2017 Aug 9;12(8):e0182666. doi: 10.1371/journal.pone.0182666. eCollection 2017. PLoS One. 2017. PMID: 28793339 Free PMC article.
-
Regulatory Genetic Networks by microRNAs: Exploring Genomic Signatures in Cervical Cancer.Biomedicines. 2025 Jun 13;13(6):1457. doi: 10.3390/biomedicines13061457. Biomedicines. 2025. PMID: 40564176 Free PMC article. Review.
-
A review on methods for predicting miRNA-mRNA regulatory modules.J Integr Bioinform. 2022 Apr 1;19(3):20200048. doi: 10.1515/jib-2020-0048. eCollection 2022 Sep 1. J Integr Bioinform. 2022. PMID: 35357793 Free PMC article. Review.
-
MicroRNA heterogeneity in melanoma progression.Semin Cancer Biol. 2019 Dec;59:208-220. doi: 10.1016/j.semcancer.2019.05.021. Epub 2019 Jun 1. Semin Cancer Biol. 2019. PMID: 31163254 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

