Arabidopsis Motif Scanner
- PMID: 26817596
- PMCID: PMC4728824
- DOI: 10.1186/s12859-016-0896-x
Arabidopsis Motif Scanner
Abstract
Background: The major mechanism driving cellular differentiation and organism development is the regulation of gene expression. Cis-acting enhancers and silencers have key roles in controlling gene transcription. The genomic era allowed the transition from single gene analysis to the investigation of full transcriptomes. This transition increased the complexity of the analyses and the difficulty in the interpretation of the results. In this context, there is demand for new tools aimed at the creation of gene networks that can facilitate the interpretation of Next Generation Sequencing (NGS) data.
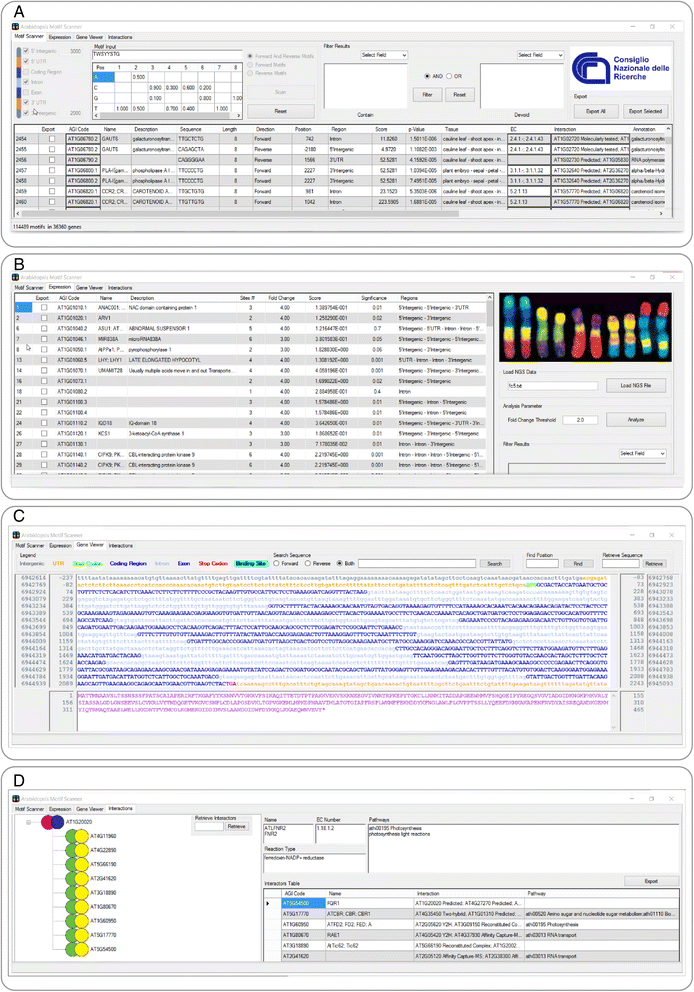
Results: Arabidopsis Motif Scanner (AMS) is a Windows application that runs on local computers. It was developed to build gene networks by identifying the positions of cis-regulatory elements in the model plant Arabidopsis thaliana and by providing an easy interface to assess and evaluate gene relationships. Its major innovative feature is to combine the cis-regulatory element positions, NGS and DNA Chip Arrays expression data, Arabidopsis annotations and gene interactions for the identification of gene networks regulated by transcription factors. In studies focused on transcription factors function, the software uses the expression data and binding site motifs in the regulative gene regions to predict direct target genes. Additionally, AMS utilizes DNA-protein and protein-protein interaction data to facilitate the identification of the metabolic pathways regulated by the transcription factor of interest.
Conclusions: Arabidopsis Motif Scanner is a new tool that helps researchers to unravel gene relations and functions. In fact, it facilitates studies focused on the effects and the impact that transcription factors have on the transcriptome by correlating the position of cis-acting elements, gene expression data and interactions.
Figures
References
-
- Susumo O. So much “junk” DNA in our genome. Smith HH, editor Evolution of genetic systems New York: Gordon and Breach 1972:366–370.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources