Oscillating behavior of Clostridium difficile Min proteins in Bacillus subtilis
- PMID: 26817670
- PMCID: PMC4905992
- DOI: 10.1002/mbo3.337
Oscillating behavior of Clostridium difficile Min proteins in Bacillus subtilis
Abstract
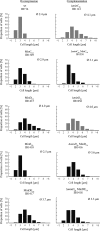
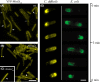
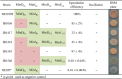
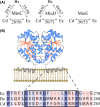
In rod-shaped bacteria, the proper placement of the division septum at the midcell relies, at least partially, on the proteins of the Min system as an inhibitor of cell division. The main principle of Min system function involves the formation of an inhibitor gradient along the cell axis; however, the establishment of this gradient differs between two well-studied gram-negative and gram-positive bacteria. While in gram-negative Escherichia coli, the Min system undergoes pole-to-pole oscillation, in gram-positive Bacillus subtilis, proper spatial inhibition is achieved by the preferential attraction of the Min proteins to the cell poles. Nevertheless, when E.coli Min proteins are inserted into B.subtilis cells, they still oscillate, which negatively affects asymmetric septation during sporulation in this organism. Interestingly, homologs of both Min systems were found to be present in various combinations in the genomes of anaerobic and endospore-forming Clostridia, including the pathogenic Clostridium difficile. Here, we have investigated the localization and behavior of C.difficile Min protein homologs and showed that MinDE proteins of C.difficile can oscillate when expressed together in B.subtilis cells. We have also investigated the effects of this oscillation on B.subtilis sporulation, and observed decreased sporulation efficiency in strains harboring the MinDE genes. Additionally, we have evaluated the effects of C.difficile Min protein expression on vegetative division in this heterologous host.
Keywords: Bacillus subtilis; Clostridium difficile; Min system oscillation; bacterial cell division; sporulation.
© 2016 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures

Similar articles
-
An oscillating Min system in Bacillus subtilis influences asymmetrical septation during sporulation.Microbiology (Reading). 2012 Aug;158(Pt 8):1972-1981. doi: 10.1099/mic.0.059295-0. Epub 2012 May 24. Microbiology (Reading). 2012. PMID: 22628484 Free PMC article.
-
Clostridial DivIVA and MinD interact in the absence of MinJ.Anaerobe. 2018 Apr;50:22-31. doi: 10.1016/j.anaerobe.2018.01.013. Epub 2018 Jan 31. Anaerobe. 2018. PMID: 29408597
-
Asymmetric cell division during Bacillus subtilis sporulation.Future Microbiol. 2019 Mar;14:353-363. doi: 10.2217/fmb-2018-0338. Epub 2019 Mar 11. Future Microbiol. 2019. PMID: 30855188
-
Clostridium difficile spore biology: sporulation, germination, and spore structural proteins.Trends Microbiol. 2014 Jul;22(7):406-16. doi: 10.1016/j.tim.2014.04.003. Epub 2014 May 7. Trends Microbiol. 2014. PMID: 24814671 Free PMC article. Review.
-
A mother cell-to-forespore channel: current understanding and future challenges.FEMS Microbiol Lett. 2014 Sep;358(2):129-36. doi: 10.1111/1574-6968.12554. Epub 2014 Aug 28. FEMS Microbiol Lett. 2014. PMID: 25105965 Review.
Cited by
-
A Strongly Fluorescing Anaerobic Reporter and Protein-Tagging System for Clostridium Organisms Based on the Fluorescence-Activating and Absorption-Shifting Tag Protein (FAST).Appl Environ Microbiol. 2019 Jul 1;85(14):e00622-19. doi: 10.1128/AEM.00622-19. Print 2019 Jul 15. Appl Environ Microbiol. 2019. PMID: 31076434 Free PMC article.
-
Clostridioides difficile Sporulation.Adv Exp Med Biol. 2024;1435:273-314. doi: 10.1007/978-3-031-42108-2_13. Adv Exp Med Biol. 2024. PMID: 38175480 Review.
-
Multistability and dynamic transitions of intracellular Min protein patterns.Mol Syst Biol. 2016 Jun 8;12(6):873. doi: 10.15252/msb.20156724. Mol Syst Biol. 2016. PMID: 27279643 Free PMC article.
-
The Min-protein oscillations in Escherichia coli: an example of self-organized cellular protein waves.Philos Trans R Soc Lond B Biol Sci. 2018 May 26;373(1747):20170111. doi: 10.1098/rstb.2017.0111. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 29632263 Free PMC article. Review.
-
Xanthomonas citri MinC Oscillates from Pole to Pole to Ensure Proper Cell Division and Shape.Front Microbiol. 2017 Jul 19;8:1352. doi: 10.3389/fmicb.2017.01352. eCollection 2017. Front Microbiol. 2017. PMID: 28769912 Free PMC article.
References
-
- Adams, D. W. , and Errington J.. 2009. Bacterial cell division, assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653. - PubMed
-
- Ausubel, F. M. , Brent R., Kingston R. E., Moore D. O., Seidmann J. S., Smith J., et al. 1987. Current Protocols in Molecular Biology. Wiley, New York.
-
- Barák, I. , and Wilkinson A. J.. 2007. Division site recognition in Escherichia coli and Bacillus subtilis . FEMS Microbiol. Rev. 31:311–326. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

