Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista
- PMID: 26817772
- PMCID: PMC4795036
- DOI: 10.1098/rspb.2015.2802
Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista
Abstract
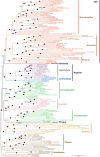
Assembling the global eukaryotic tree of life has long been a major effort of Biology. In recent years, pushed by the new availability of genome-scale data for microbial eukaryotes, it has become possible to revisit many evolutionary enigmas. However, some of the most ancient nodes, which are essential for inferring a stable tree, have remained highly controversial. Among other reasons, the lack of adequate genomic datasets for key taxa has prevented the robust reconstruction of early diversification events. In this context, the centrohelid heliozoans are particularly relevant for reconstructing the tree of eukaryotes because they represent one of the last substantial groups that was missing large and diverse genomic data. Here, we filled this gap by sequencing high-quality transcriptomes for four centrohelid lineages, each corresponding to a different family. Combining these new data with a broad eukaryotic sampling, we produced a gene-rich taxon-rich phylogenomic dataset that enabled us to refine the structure of the tree. Specifically, we show that (i) centrohelids relate to haptophytes, confirming Haptista; (ii) Haptista relates to SAR; (iii) Cryptista share strong affinity with Archaeplastida; and (iv) Haptista + SAR is sister to Cryptista + Archaeplastida. The implications of this topology are discussed in the broader context of plastid evolution.
Keywords: centrohelids; eukaryotes; phylogenomics; plastid evolution; tree of life.
© 2016 The Author(s).
Figures


References
-
- Mikrjukov KA. 2000. System and phylogeny of Heliozoa: should this taxon exist in modern systems of protists? Zool Z. 79, 883–897.
-
- Cavalier-Smith T. 2003. Protist phylogeny and the high-level classification of Protozoa. Eur. J. Protistol. 39, 338–348. ( 10.1078/0932-4739-00002) - DOI
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
