Scorpion Potassium Channel-blocking Defensin Highlights a Functional Link with Neurotoxin
- PMID: 26817841
- PMCID: PMC4807291
- DOI: 10.1074/jbc.M115.680611
Scorpion Potassium Channel-blocking Defensin Highlights a Functional Link with Neurotoxin
Abstract
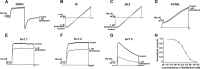
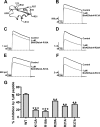
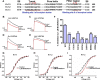
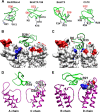
The structural similarity between defensins and scorpion neurotoxins suggests that they might have evolved from a common ancestor. However, there is no direct experimental evidence demonstrating a functional link between scorpion neurotoxins and defensins. The scorpion defensin BmKDfsin4 from Mesobuthus martensiiKarsch contains 37 amino acid residues and a conserved cystine-stabilized α/β structural fold. The recombinant BmKDfsin4, a classical defensin, has been found to have inhibitory activity against Gram-positive bacteria such as Staphylococcus aureus, Bacillus subtilis, and Micrococcus luteusas well as methicillin-resistant Staphylococcus aureus Interestingly, electrophysiological experiments showed that BmKDfsin4,like scorpion potassium channel neurotoxins, could effectively inhibit Kv1.1, Kv1.2, and Kv1.3 channel currents, and its IC50value for the Kv1.3 channel was 510.2 nm Similar to the structure-function relationships of classical scorpion potassium channel-blocking toxins, basic residues (Lys-13 and Arg-19) of BmKDfsin4 play critical roles in peptide-Kv1.3 channel interactions. Furthermore, mutagenesis and electrophysiological experiments demonstrated that the channel extracellular pore region is the binding site of BmKDfsin4, indicating that BmKDfsin4 adopts the same mechanism for blocking potassium channel currents as classical scorpion toxins. Taken together, our work identifies scorpion BmKDfsin4 as the first invertebrate defensin to block potassium channels. These findings not only demonstrate that defensins from invertebrate animals are a novel type of potassium channel blockers but also provide evidence of a functional link between defensins and neurotoxins.
Keywords: bacteria; defensin; neurotoxin; potassium channel; structure-function.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Dunlop J. A., Kamenz C., and Scholtz G. (2007) Reinterpreting the morphology of the Jurassic scorpion Liassoscorpionides. Arthropod Struct. Dev. 36, 245–252 - PubMed
-
- Fry B. G., Roelants K., Champagne D. E., Scheib H., Tyndall J. D., King G. F., Nevalainen T. J., Norman J. A., Lewis R. J., Norton R. S., Renjifo C., and de la Vega R. C. (2009) The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 10, 483–511 - PubMed
-
- Batista C. V., D'Suze G., Gómez-Lagunas F., Zamudio F. Z., Encarnación S., Sevcik C., and Possani L. D. (2006) Proteomic analysis of Tityus discrepans scorpion venom and amino acid sequence of novel toxins. Proteomics 6, 3718–3727 - PubMed
-
- de Oliveira U. C., Candido D. M., Coronado Dorce V. A., and Junqueira-de-Azevedo I. L. (2015) The transcriptome recipe for the venom cocktail of Tityus bahiensis scorpion. Toxicon 95, 52–61 - PubMed
-
- Xu X., Duan Z., Di Z., He Y., Li J., Li Z., Xie C., Zeng X., Cao Z., Wu Y., Liang S., and Li W. (2014) Proteomic analysis of the venom from the scorpion Mesobuthus martensii. J. Proteomics 106, 162–180 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

