Editing of Cellular Self-RNAs by Adenosine Deaminase ADAR1 Suppresses Innate Immune Stress Responses
- PMID: 26817845
- PMCID: PMC4813567
- DOI: 10.1074/jbc.M115.709014
Editing of Cellular Self-RNAs by Adenosine Deaminase ADAR1 Suppresses Innate Immune Stress Responses
Abstract
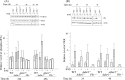
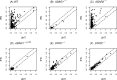
Adenosine deaminases acting on double-stranded RNA (ADARs) catalyze the deamination of adenosine (A) to produce inosine (I) in double-stranded (ds) RNA structures, a process known as A-to-I RNA editing. dsRNA is an important trigger of innate immune responses, including interferon (IFN) production and action. We examined the role of A-to-I RNA editing by two ADARs, ADAR1 and ADAR2, in the sensing of self-RNA in the absence of pathogen infection, leading to activation of IFN-induced, RNA-mediated responses in mouse embryo fibroblasts. IFN treatment of Adar1(-/-) cells lacking both the p110 constitutive and p150 IFN-inducible ADAR1 proteins induced formation of stress granules, whereas neither wild-type (WT) nor Adar2(-/-) cells displayed a comparable stress granule response following IFN treatment. Phosphorylation of protein synthesis initiation factor eIF2α at serine 51 was increased in IFN-treated Adar1(-/-) cells but not in either WT or Adar2(-/-) cells following IFN treatment. Analysis by deep sequencing of mouse exonic loci containing A-to-I-editing sites revealed that the majority of editing in mouse embryo fibroblasts was carried out by ADAR1. IFN treatment increased editing in both WT and Adar2(-/-) cells but not in either Adar1(-/-) or Adar1(-/-) (p150) cells or Stat1(-/-) or Stat2(-/-) cells. Hyper-edited sites found in predicted duplex structures showed strand bias of editing for some RNAs. These results implicate ADAR1 p150 as the major A-to-I editor in mouse embryo fibroblasts, acting as a feedback suppressor of innate immune responses otherwise triggered by self-RNAs possessing regions of double-stranded character.
Keywords: RNA editing; adenosine deaminase acting on RNA (ADAR); innate immunity; interferon; protein kinase RNA-activated (PKR); stress granule.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
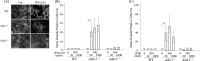

Similar articles
-
An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1).J Interferon Cytokine Res. 2014 Jun;34(6):437-46. doi: 10.1089/jir.2014.0001. J Interferon Cytokine Res. 2014. PMID: 24905200 Free PMC article. Review.
-
Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses.J Biol Chem. 2019 Feb 1;294(5):1710-1720. doi: 10.1074/jbc.TM118.004166. J Biol Chem. 2019. PMID: 30710018 Free PMC article. Review.
-
Adenosine Deaminases Acting on RNA (ADARs) and Viral Infections.Annu Rev Virol. 2021 Sep 29;8(1):239-264. doi: 10.1146/annurev-virology-091919-065320. Epub 2021 Apr 21. Annu Rev Virol. 2021. PMID: 33882257
-
Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1p150.PLoS Biol. 2018 Nov 29;16(11):e2006577. doi: 10.1371/journal.pbio.2006577. eCollection 2018 Nov. PLoS Biol. 2018. PMID: 30496178 Free PMC article.
-
Chimeric double-stranded RNA-specific adenosine deaminase ADAR1 proteins reveal functional selectivity of double-stranded RNA-binding domains from ADAR1 and protein kinase PKR.Proc Natl Acad Sci U S A. 2000 Nov 7;97(23):12541-6. doi: 10.1073/pnas.97.23.12541. Proc Natl Acad Sci U S A. 2000. PMID: 11070079 Free PMC article.
Cited by
-
The majority of A-to-I RNA editing is not required for mammalian homeostasis.Genome Biol. 2019 Dec 9;20(1):268. doi: 10.1186/s13059-019-1873-2. Genome Biol. 2019. PMID: 31815657 Free PMC article.
-
Z-DNA and Z-RNA: Methods-Past and Future.Methods Mol Biol. 2023;2651:295-329. doi: 10.1007/978-1-0716-3084-6_21. Methods Mol Biol. 2023. PMID: 36892776
-
IFN-I inducible miR-3614-5p targets ADAR1 isoforms and fine tunes innate immune activation.Front Immunol. 2022 Jul 22;13:939907. doi: 10.3389/fimmu.2022.939907. eCollection 2022. Front Immunol. 2022. PMID: 35935998 Free PMC article.
-
Adenosine-to-inosine RNA editing in the immune system: friend or foe?Cell Mol Life Sci. 2020 Aug;77(15):2931-2948. doi: 10.1007/s00018-020-03466-2. Epub 2020 Jan 29. Cell Mol Life Sci. 2020. PMID: 31996954 Free PMC article. Review.
-
Protein kinase R and the integrated stress response drive immunopathology caused by mutations in the RNA deaminase ADAR1.Immunity. 2021 Sep 14;54(9):1948-1960.e5. doi: 10.1016/j.immuni.2021.07.001. Epub 2021 Aug 2. Immunity. 2021. PMID: 34343497 Free PMC article.
References
-
- Holcik M., and Sonenberg N. (2005) Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327 - PubMed
-
- Samuel C. E. (1993) The eIF-2α protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268, 7603–7606 - PubMed
-
- Wek R. C., Jiang H. Y., and Anthony T. G. (2006) Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 - PubMed
-
- Anderson P., and Kedersha N. (2009) RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430–436 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

