Identification of pH-sensing Sites in the Light Harvesting Complex Stress-related 3 Protein Essential for Triggering Non-photochemical Quenching in Chlamydomonas reinhardtii
- PMID: 26817847
- PMCID: PMC4817166
- DOI: 10.1074/jbc.M115.704601
Identification of pH-sensing Sites in the Light Harvesting Complex Stress-related 3 Protein Essential for Triggering Non-photochemical Quenching in Chlamydomonas reinhardtii
Abstract
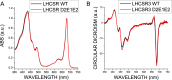
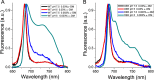
Light harvesting complex stress-related 3 (LHCSR3) is the protein essential for photoprotective excess energy dissipation (non-photochemical quenching, NPQ) in the model green algaChlamydomonas reinhardtii Activation of NPQ requires low pH in the thylakoid lumen, which is induced in excess light conditions and sensed by lumen-exposed acidic residues. In this work we have used site-specific mutagenesisin vivoandin vitrofor identification of the residues in LHCSR3 that are responsible for sensing lumen pH. Lumen-exposed protonatable residues, aspartate and glutamate, were mutated to asparagine and glutamine, respectively. By expression in a mutant lacking all LHCSR isoforms, residues Asp(117), Glu(221), and Glu(224)were shown to be essential for LHCSR3-dependent NPQ induction inC. reinhardtii Analysis of recombinant proteins carrying the same mutations refoldedin vitrowith pigments showed that the capacity of responding to low pH by decreasing the fluorescence lifetime, present in the wild-type protein, was lost. Consistent with a role in pH sensing, the mutations led to a substantial reduction in binding the NPQ inhibitor dicyclohexylcarbodiimide.
Keywords: fluorescence; non-photochemical quenching; photoprotection; photosynthesis; photosynthetic pigment; photosystem II; plant biochemistry.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii.Proc Natl Acad Sci U S A. 2013 Jun 11;110(24):10016-21. doi: 10.1073/pnas.1222606110. Epub 2013 May 28. Proc Natl Acad Sci U S A. 2013. PMID: 23716695 Free PMC article.
-
Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii.PLoS Biol. 2011 Jan 18;9(1):e1000577. doi: 10.1371/journal.pbio.1000577. PLoS Biol. 2011. PMID: 21267060 Free PMC article.
-
LHCSR3 is a nonphotochemical quencher of both photosystems in Chlamydomonas reinhardtii.Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):4212-4217. doi: 10.1073/pnas.1809812116. Epub 2019 Feb 19. Proc Natl Acad Sci U S A. 2019. PMID: 30782831 Free PMC article.
-
Novel insights into the function of LHCSR3 in Chlamydomonas reinhardtii.Plant Signal Behav. 2015;10(12):e1058462. doi: 10.1080/15592324.2015.1058462. Plant Signal Behav. 2015. PMID: 26237677 Free PMC article. Review.
-
Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis.Curr Opin Plant Biol. 2013 Jun;16(3):307-14. doi: 10.1016/j.pbi.2013.03.011. Epub 2013 Apr 11. Curr Opin Plant Biol. 2013. PMID: 23583332 Review.
Cited by
-
The relationship between photosystem II regulation and light-dependent hydrogen production by microalgae.Biophys Rev. 2022 Jul 15;14(4):893-904. doi: 10.1007/s12551-022-00977-z. eCollection 2022 Aug. Biophys Rev. 2022. PMID: 36124275 Free PMC article. Review.
-
Transcriptional regulation of photoprotection in dark-to-light transition-More than just a matter of excess light energy.Sci Adv. 2022 Jun 3;8(22):eabn1832. doi: 10.1126/sciadv.abn1832. Epub 2022 Jun 3. Sci Adv. 2022. PMID: 35658034 Free PMC article.
-
LHCSR1-dependent fluorescence quenching is mediated by excitation energy transfer from LHCII to photosystem I in Chlamydomonas reinhardtii.Proc Natl Acad Sci U S A. 2018 Apr 3;115(14):3722-3727. doi: 10.1073/pnas.1720574115. Epub 2018 Mar 19. Proc Natl Acad Sci U S A. 2018. PMID: 29555769 Free PMC article.
-
Potential and Challenges of Improving Photosynthesis in Algae.Plants (Basel). 2020 Jan 3;9(1):67. doi: 10.3390/plants9010067. Plants (Basel). 2020. PMID: 31947868 Free PMC article. Review.
-
Concentration Effect on Quenching of Chlorophyll a Fluorescence by All-Trans-β-Carotene in Photosynthesis.Molecules. 2017 Sep 21;22(10):1585. doi: 10.3390/molecules22101585. Molecules. 2017. PMID: 28934156 Free PMC article.
References
-
- Niyogi K. K. (1999) Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359 - PubMed
-
- Rees D., Noctor G., Ruban A. V., Crofts J., Young A., and Horton P. (1992) pH dependent chlorophyll fluorescence quenching in spinach thylakoids from light treated or dark adapted leaves. Photosynth. Res. 31, 11–19 - PubMed
-
- de Bianchi S., Ballottari M., Dall'osto L., and Bassi R. (2010) Regulation of plant light harvesting by thermal dissipation of excess energy. Biochem. Soc. Trans. 38, 651–660 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

