CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients
- PMID: 26818188
- PMCID: PMC4730182
- DOI: 10.1038/srep20070
CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients
Erratum in
-
Corrigendum: CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients.Sci Rep. 2017 Jan 19;7:40272. doi: 10.1038/srep40272. Sci Rep. 2017. PMID: 28102301 Free PMC article. No abstract available.
Abstract
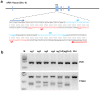
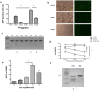
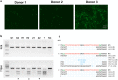
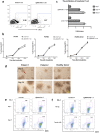
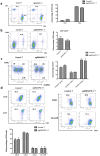
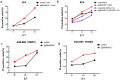
Strategies that enhance the function of T cells are critical for immunotherapy. One negative regulator of T-cell activity is ligand PD-L1, which is expressed on dentritic cells (DCs) or some tumor cells, and functions through binding of programmed death-1 (PD-1) receptor on activated T cells. Here we described for the first time a non-viral mediated approach to reprogram primary human T cells by disruption of PD-1. We showed that the gene knockout of PD-1 by electroporation of plasmids encoding sgRNA and Cas9 was technically feasible. The disruption of inhibitory checkpoint gene PD-1 resulted in significant reduction of PD-1 expression but didn't affect the viability of primary human T cells during the prolonged in vitro culture. Cellular immune response of the gene modified T cells was characterized by up-regulated IFN-γ production and enhanced cytotoxicity. These results suggest that we have demonstrated an approach for efficient checkpoint inhibitor disruption in T cells, providing a new strategy for targeting checkpoint inhibitors, which could potentialy be useful to improve the efficacy of T-cell based adoptive therapies.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

