Transcriptome sequencing based annotation and homologous evidence based scaffolding of Anguilla japonica draft genome
- PMID: 26818233
- PMCID: PMC4895481
- DOI: 10.1186/s12864-015-2306-6
Transcriptome sequencing based annotation and homologous evidence based scaffolding of Anguilla japonica draft genome
Abstract
Background: Anguilla japonica (Japanese eel) is currently one of the most important research subjects in eastern Asia aquaculture. Enigmatic life cycle of the organism makes study of artificial reproduction extremely limited. Henceforth genomic and transcriptomic resources of eels are urgently needed to help solving the problems surrounding this organism across multiple fields. We hereby provide a reconstructed transcriptome from deep sequencing of juvenile (glass eels) whole body samples. The provided expressed sequence tags were used to annotate the currently available draft genome sequence. Homologous information derived from the annotation result was applied to improve the group of scaffolds into available linkage groups.
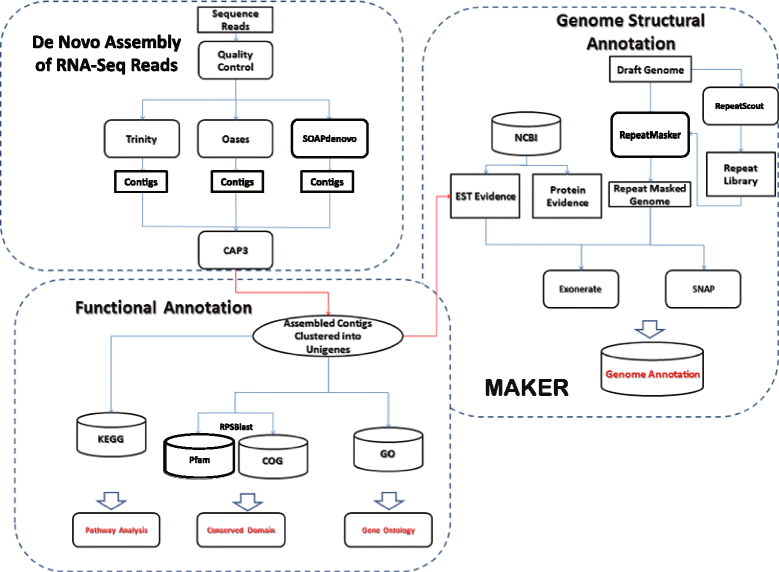
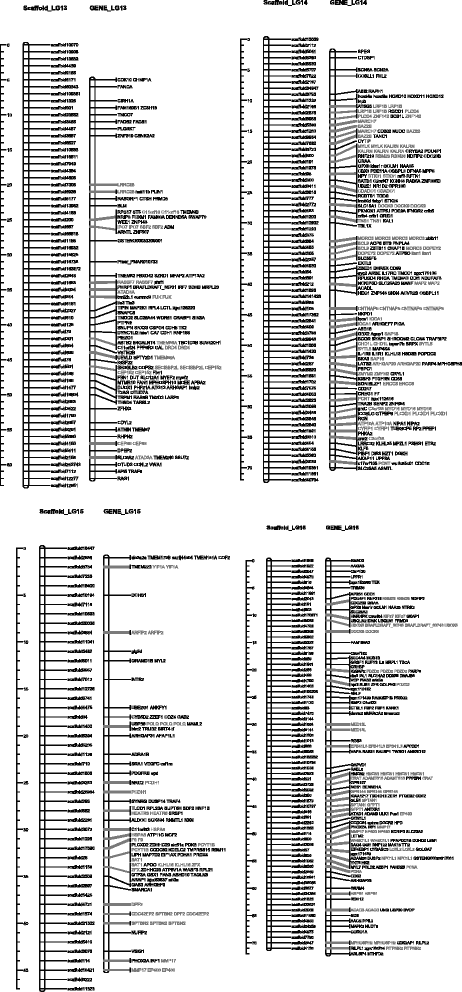
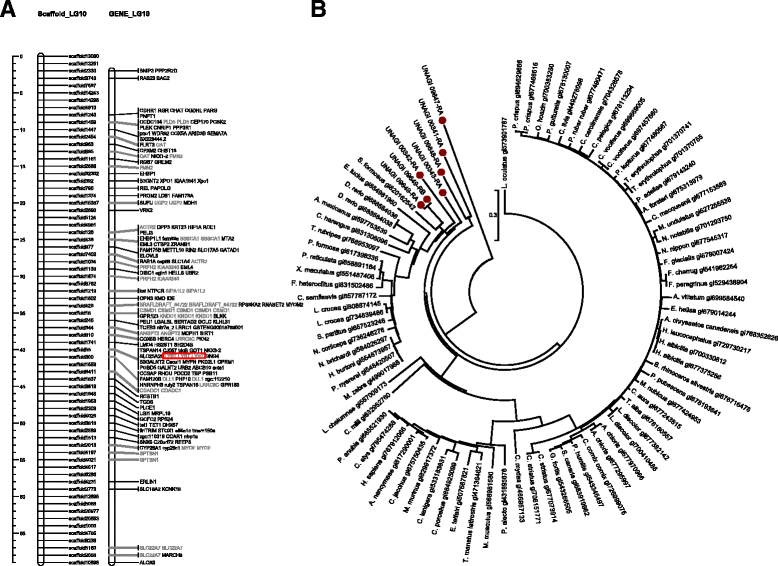
Results: With the transcriptome sequence data combined with publicly available expressed sequence tags evidences, 18,121 genes were structurally and functionally annotated on the draft genome. Among them, 3,921 genes were located in the 19 linkage groups. 137 scaffolds covering 13 million bases were grouped into the linkage groups in additional to the original partial linkage groups, increasing the linkage group coverage from 13 to 14%.
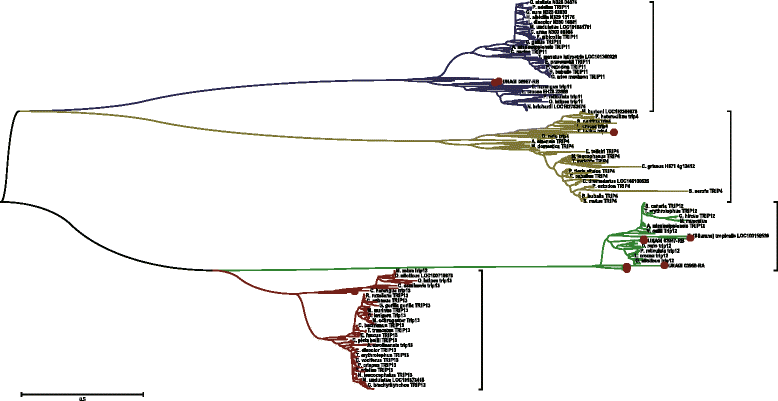
Conclusions: This annotation provide information of the coding regions of the genes supported by transcriptome based evidence. The derived homologous evidences pave the way for phylogenetic analysis of important genetic traits and the improvement of the genome assembly.
Figures







Similar articles
-
Identification of genetic linkage group 1-linked sequences in Japanese eel (Anguilla japonica) by single chromosome sorting and sequencing.PLoS One. 2018 May 8;13(5):e0197040. doi: 10.1371/journal.pone.0197040. eCollection 2018. PLoS One. 2018. PMID: 29738551 Free PMC article.
-
A ddRAD-based genetic map and its integration with the genome assembly of Japanese eel (Anguilla japonica) provides insights into genome evolution after the teleost-specific genome duplication.BMC Genomics. 2014 Mar 26;15:233. doi: 10.1186/1471-2164-15-233. BMC Genomics. 2014. PMID: 24669946 Free PMC article.
-
Genetic parameters and quantitative trait loci analysis associated with body size and timing at metamorphosis into glass eels in captive-bred Japanese eels (Anguilla japonica).PLoS One. 2018 Aug 29;13(8):e0201784. doi: 10.1371/journal.pone.0201784. eCollection 2018. PLoS One. 2018. PMID: 30157280 Free PMC article.
-
First draft genome sequence of the Japanese eel, Anguilla japonica.Gene. 2012 Dec 15;511(2):195-201. doi: 10.1016/j.gene.2012.09.064. Epub 2012 Sep 29. Gene. 2012. PMID: 23026207
-
Draft genome of the American Eel (Anguilla rostrata).Mol Ecol Resour. 2017 Jul;17(4):806-811. doi: 10.1111/1755-0998.12608. Epub 2016 Nov 14. Mol Ecol Resour. 2017. PMID: 27754597
Cited by
-
Physiological mechanism of osmoregulatory adaptation in anguillid eels.Fish Physiol Biochem. 2018 Apr;44(2):423-433. doi: 10.1007/s10695-018-0464-6. Epub 2018 Jan 17. Fish Physiol Biochem. 2018. PMID: 29344774 Free PMC article. Review.
-
A Chromosome-level assembly of the Japanese eel genome, insights into gene duplication and chromosomal reorganization.Gigascience. 2022 Dec 8;11:giac120. doi: 10.1093/gigascience/giac120. Gigascience. 2022. PMID: 36480030 Free PMC article.
References
-
- Tsukamoto K, Aoyama J, Miller MJ. Migration, speciation, and the evolution of diadromy in anguillid eels. Can J Fish Aquat Sci. 2002;59(12):1989–1998. doi: 10.1139/f02-165. - DOI
-
- Nomura K, Ozaki A, Morishima K, Yoshikawa Y, Tanaka H, Unuma T, et al. A genetic linkage map of the Japanese eel (Anguilla japonica) based on AFLP and microsatellite markers. Aquaculture. 2011;310(3–4):329–342. doi: 10.1016/j.aquaculture.2010.11.006. - DOI
-
- Coppe A, Pujolar JM, Maes GE, Larsen PF, Hansen MM, Bernatchez L, et al. Sequencing, de novo annotation and analysis of the first Anguilla anguilla transcriptome: EeelBase opens new perspectives for the study of the critically endangered european eel. BMC Genomics. 2010;11:635. doi: 10.1186/1471-2164-11-635. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

