Inference of domain-disease associations from domain-protein, protein-disease and disease-disease relationships
- PMID: 26818594
- PMCID: PMC4895779
- DOI: 10.1186/s12918-015-0247-y
Inference of domain-disease associations from domain-protein, protein-disease and disease-disease relationships
Abstract
Background: Protein domains can be viewed as portable units of biological function that defines the functional properties of proteins. Therefore, if a protein is associated with a disease, protein domains might also be associated and define disease endophenotypes. However, knowledge about such domain-disease relationships is rarely available. Thus, identification of domains associated with human diseases would greatly improve our understanding of the mechanism of human complex diseases and further improve the prevention, diagnosis and treatment of these diseases.
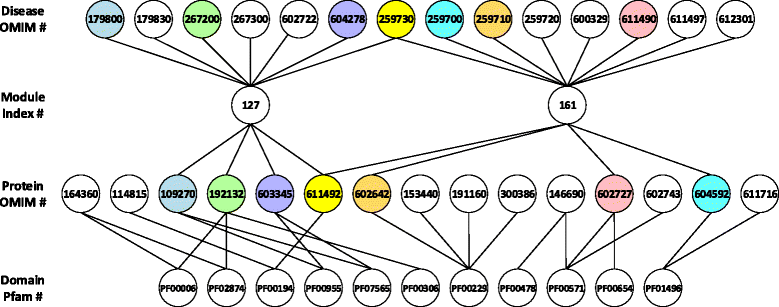
Methods: Based on phenotypic similarities among diseases, we first group diseases into overlapping modules. We then develop a framework to infer associations between domains and diseases through known relationships between diseases and modules, domains and proteins, as well as proteins and disease modules. Different methods including Association, Maximum likelihood estimation (MLE), Domain-disease pair exclusion analysis (DPEA), Bayesian, and Parsimonious explanation (PE) approaches are developed to predict domain-disease associations.
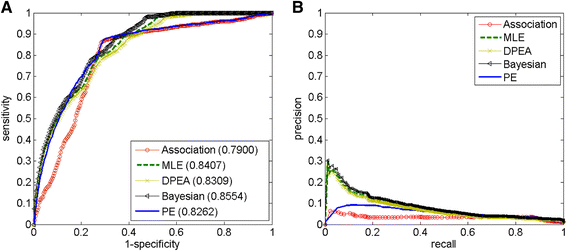
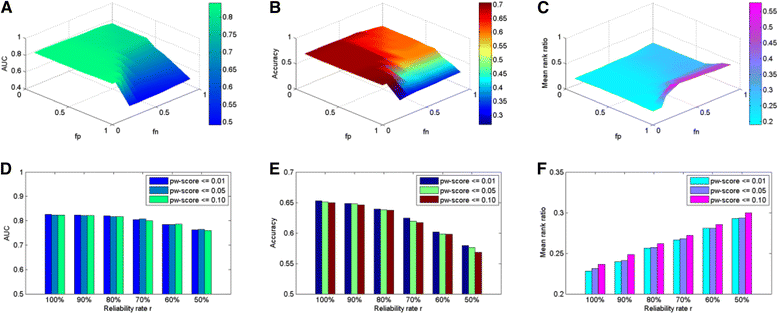
Results: We demonstrate the effectiveness of all the five approaches via a series of validation experiments, and show the robustness of the MLE, Bayesian and PE approaches to the involved parameters. We also study the effects of disease modularization in inferring novel domain-disease associations. Through validation, the AUC (Area Under the operating characteristic Curve) scores for Bayesian, MLE, DPEA, PE, and Association approaches are 0.86, 0.84, 0.83, 0.83 and 0.79, respectively, indicating the usefulness of these approaches for predicting domain-disease relationships. Finally, we choose the Bayesian approach to infer domains associated with two common diseases, Crohn's disease and type 2 diabetes.
Conclusions: The Bayesian approach has the best performance for the inference of domain-disease relationships. The predicted landscape between domains and diseases provides a more detailed view about the disease mechanisms.
Figures

Similar articles
-
Protein structural domain-disease association prediction based on heterogeneous networks.BMC Genomics. 2025 Apr 10;23(Suppl 6):869. doi: 10.1186/s12864-024-11117-0. BMC Genomics. 2025. PMID: 40211147 Free PMC article.
-
DomainRBF: a Bayesian regression approach to the prioritization of candidate domains for complex diseases.BMC Syst Biol. 2011 Apr 19;5:55. doi: 10.1186/1752-0509-5-55. BMC Syst Biol. 2011. PMID: 21504591 Free PMC article.
-
Evaluation of different domain-based methods in protein interaction prediction.Biochem Biophys Res Commun. 2009 Dec 18;390(3):357-62. doi: 10.1016/j.bbrc.2009.09.130. Epub 2009 Oct 2. Biochem Biophys Res Commun. 2009. PMID: 19800868 Review.
-
Prioritisation of associations between protein domains and complex diseases using domain-domain interaction networks.IET Syst Biol. 2010 May;4(3):212-22. doi: 10.1049/iet-syb.2009.0037. IET Syst Biol. 2010. PMID: 20500001
-
Temporal abstraction and temporal Bayesian networks in clinical domains: a survey.Artif Intell Med. 2014 Mar;60(3):133-49. doi: 10.1016/j.artmed.2013.12.007. Epub 2014 Jan 17. Artif Intell Med. 2014. PMID: 24529699 Review.
Cited by
-
Mapping OMIM Disease-Related Variations on Protein Domains Reveals an Association Among Variation Type, Pfam Models, and Disease Classes.Front Mol Biosci. 2021 May 7;8:617016. doi: 10.3389/fmolb.2021.617016. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026820 Free PMC article.
-
Protein structural domain-disease association prediction based on heterogeneous networks.BMC Genomics. 2025 Apr 10;23(Suppl 6):869. doi: 10.1186/s12864-024-11117-0. BMC Genomics. 2025. PMID: 40211147 Free PMC article.
-
Pathogenic variation types in human genes relate to diseases through Pfam and InterPro mapping.Front Mol Biosci. 2022 Sep 16;9:966927. doi: 10.3389/fmolb.2022.966927. eCollection 2022. Front Mol Biosci. 2022. PMID: 36188216 Free PMC article.
-
DapBCH: a disease association prediction model Based on Cross-species and Heterogeneous graph embedding.Front Genet. 2023 Sep 22;14:1222346. doi: 10.3389/fgene.2023.1222346. eCollection 2023. Front Genet. 2023. PMID: 37811150 Free PMC article.
-
CCDC66 frameshift variant associated with a new form of early-onset progressive retinal atrophy in Portuguese Water Dogs.Sci Rep. 2020 Dec 3;10(1):21162. doi: 10.1038/s41598-020-77980-5. Sci Rep. 2020. PMID: 33273526 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

