SEUSS Integrates Gibberellin Signaling with Transcriptional Inputs from the SHR-SCR-SCL3 Module to Regulate Middle Cortex Formation in the Arabidopsis Root
- PMID: 26818732
- PMCID: PMC4775121
- DOI: 10.1104/pp.15.01501
SEUSS Integrates Gibberellin Signaling with Transcriptional Inputs from the SHR-SCR-SCL3 Module to Regulate Middle Cortex Formation in the Arabidopsis Root
Abstract
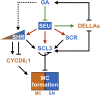
A decade of studies on middle cortex (MC) formation in the root endodermis of Arabidopsis (Arabidopsis thaliana) have revealed a complex regulatory network that is orchestrated by several GRAS family transcription factors, including SHORT-ROOT (SHR), SCARECROW (SCR), and SCARECROW-LIKE3 (SCL3). However, how their functions are regulated remains obscure. Here we show that mutations in the SEUSS (SEU) gene led to a higher frequency of MC formation. seu mutants had strongly reduced expression of SHR, SCR, and SCL3, suggesting that SEU positively regulates these genes. Our results further indicate that SEU physically associates with upstream regulatory sequences of SHR, SCR, and SCL3; and that SEU has distinct genetic interactions with these genes in the control of MC formation, with SCL3 being epistatic to SEU. Similar to SCL3, SEU was repressed by the phytohormone GA and induced by the GA biosynthesis inhibitor paclobutrazol, suggesting that SEU acts downstream of GA signaling to regulate MC formation. Consistently, we found that SEU mediates the regulation of SCL3 by GA signaling. Together, our study identifies SEU as a new critical player that integrates GA signaling with transcriptional inputs from the SHR-SCR-SCL3 module to regulate MC formation in the Arabidopsis root.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures




Similar articles
-
SCARECROW-LIKE3 regulates the transcription of gibberellin-related genes by acting as a transcriptional co-repressor of GAI-ASSOCIATED FACTOR1.Plant Mol Biol. 2021 Mar;105(4-5):463-482. doi: 10.1007/s11103-020-01101-z. Epub 2021 Jan 21. Plant Mol Biol. 2021. PMID: 33474657
-
DELLA and SCL3 balance gibberellin feedback regulation by utilizing INDETERMINATE DOMAIN proteins as transcriptional scaffolds.Plant Signal Behav. 2014;9(9):e29726. doi: 10.4161/psb.29726. Plant Signal Behav. 2014. PMID: 25763707 Free PMC article. Review.
-
Gene Regulation via the Combination of Transcription Factors in the INDETERMINATE DOMAIN and GRAS Families.Genes (Basel). 2020 Jun 2;11(6):613. doi: 10.3390/genes11060613. Genes (Basel). 2020. PMID: 32498388 Free PMC article.
-
SCARECROW-LIKE23 and SCARECROW jointly specify endodermal cell fate but distinctly control SHORT-ROOT movement.Plant J. 2015 Nov;84(4):773-84. doi: 10.1111/tpj.13038. Plant J. 2015. PMID: 26415082
-
Defining the Path from Stem Cells to Differentiated Tissue.Curr Top Dev Biol. 2016;116:35-43. doi: 10.1016/bs.ctdb.2015.12.002. Epub 2016 Feb 8. Curr Top Dev Biol. 2016. PMID: 26970612 Review.
Cited by
-
Root Patterning: Tuning SHORT ROOT Function Creates Diversity in Form.Front Plant Sci. 2021 Sep 30;12:745861. doi: 10.3389/fpls.2021.745861. eCollection 2021. Front Plant Sci. 2021. PMID: 34659316 Free PMC article. Review.
-
Transcriptional control of hydrogen peroxide homeostasis regulates ground tissue patterning in the Arabidopsis root.Front Plant Sci. 2023 Aug 21;14:1242211. doi: 10.3389/fpls.2023.1242211. eCollection 2023. Front Plant Sci. 2023. PMID: 37670865 Free PMC article.
-
A molecular framework controlling style morphology in Brassicaceae.Development. 2018 Mar 1;145(5):dev158105. doi: 10.1242/dev.158105. Development. 2018. PMID: 29440299 Free PMC article.
-
Control of Asymmetric Cell Divisions during Root Ground Tissue Maturation.Mol Cells. 2016 Jul;39(7):524-9. doi: 10.14348/molcells.2016.0105. Epub 2016 Jun 16. Mol Cells. 2016. PMID: 27306644 Free PMC article. Review.
-
Genome-Wide Characterization of GRAS Family and Their Potential Roles in Cold Tolerance of Cucumber (Cucumis sativus L.).Int J Mol Sci. 2020 May 29;21(11):3857. doi: 10.3390/ijms21113857. Int J Mol Sci. 2020. PMID: 32485801 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

