A Hepatitis C Virus Envelope Polymorphism Confers Resistance to Neutralization by Polyclonal Sera and Broadly Neutralizing Monoclonal Antibodies
- PMID: 26819308
- PMCID: PMC4794665
- DOI: 10.1128/JVI.02837-15
A Hepatitis C Virus Envelope Polymorphism Confers Resistance to Neutralization by Polyclonal Sera and Broadly Neutralizing Monoclonal Antibodies
Abstract
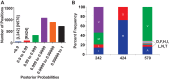
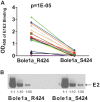
Hepatitis C virus (HCV) infection is a global health problem, with millions of chronically infected individuals at risk for cirrhosis and hepatocellular carcinoma. HCV vaccine development is vital in the effort toward disease control and eradication, an undertaking aided by an increased understanding of the mechanisms of resistance to broadly neutralizing antibodies (bNAbs). In this study, we identified HCV codons that vary deep in a phylogenetic tree of HCV sequences and showed that a polymorphism at one of these positions renders Bole1a, a computationally derived, ancestral genotype 1a HCV strain, resistant to neutralization by both polyclonal-HCV-infected plasma and multiple broadly neutralizing monoclonal antibodies with unique binding epitopes. This bNAb resistance mutation reduces replicative fitness, which may explain the persistence of both neutralization-sensitive and neutralization-resistant variants in circulating viral strains. This work identifies an important determinant of bNAb resistance in an ancestral, representative HCV genome, which may inform HCV vaccine development.
Importance: Worldwide, more than 170 million people are infected with hepatitis C virus (HCV), the leading cause of hepatocellular carcinoma and liver transplantation in the United States. Despite recent significant advances in HCV treatment, a vaccine is needed. Control of the HCV pandemic with drug treatment alone is likely to fail due to limited access to treatment, reinfections in high-risk individuals, and the potential for resistance to direct-acting antivirals (DAAs). Broadly neutralizing antibodies (bNAbs) block infection by diverse HCV variants and therefore serve as a useful guide for vaccine development, but our understanding of resistance to bNAbs is incomplete. In this report, we identify a viral polymorphism conferring resistance to neutralization by both polyclonal plasma and broadly neutralizing monoclonal antibodies, which may inform HCV vaccine development.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Broadly Neutralizing Antibodies Targeting New Sites of Vulnerability in Hepatitis C Virus E1E2.J Virol. 2019 Jun 28;93(14):e02070-18. doi: 10.1128/JVI.02070-18. Print 2019 Jul 15. J Virol. 2019. PMID: 31068427 Free PMC article.
-
Extra-epitopic hepatitis C virus polymorphisms confer resistance to broadly neutralizing antibodies by modulating binding to scavenger receptor B1.PLoS Pathog. 2017 Feb 24;13(2):e1006235. doi: 10.1371/journal.ppat.1006235. eCollection 2017 Feb. PLoS Pathog. 2017. PMID: 28235087 Free PMC article.
-
Escape of Hepatitis C Virus from Epitope I Neutralization Increases Sensitivity of Other Neutralization Epitopes.J Virol. 2018 Apr 13;92(9):e02066-17. doi: 10.1128/JVI.02066-17. Print 2018 May 1. J Virol. 2018. PMID: 29467319 Free PMC article.
-
Defining Breadth of Hepatitis C Virus Neutralization.Front Immunol. 2018 Aug 2;9:1703. doi: 10.3389/fimmu.2018.01703. eCollection 2018. Front Immunol. 2018. PMID: 30116237 Free PMC article. Review.
-
Virus-neutralizing antibodies to hepatitis C virus.J Viral Hepat. 2013 Jun;20(6):369-76. doi: 10.1111/jvh.12094. Epub 2013 Apr 4. J Viral Hepat. 2013. PMID: 23647953 Review.
Cited by
-
Mechanisms of HCV resistance to broadly neutralizing antibodies.Curr Opin Virol. 2021 Oct;50:23-29. doi: 10.1016/j.coviro.2021.07.003. Epub 2021 Jul 28. Curr Opin Virol. 2021. PMID: 34329953 Free PMC article. Review.
-
Mechanisms of Hepatitis C Virus Escape from Vaccine-Relevant Neutralizing Antibodies.Vaccines (Basel). 2021 Mar 20;9(3):291. doi: 10.3390/vaccines9030291. Vaccines (Basel). 2021. PMID: 33804732 Free PMC article. Review.
-
Broadly Neutralizing Antibodies Targeting New Sites of Vulnerability in Hepatitis C Virus E1E2.J Virol. 2019 Jun 28;93(14):e02070-18. doi: 10.1128/JVI.02070-18. Print 2019 Jul 15. J Virol. 2019. PMID: 31068427 Free PMC article.
-
HIV-antibody complexes enhance production of type I interferon by plasmacytoid dendritic cells.J Clin Invest. 2017 Dec 1;127(12):4352-4364. doi: 10.1172/JCI95375. Epub 2017 Oct 30. J Clin Invest. 2017. PMID: 29083319 Free PMC article.
-
Synergistic anti-HCV broadly neutralizing human monoclonal antibodies with independent mechanisms.Proc Natl Acad Sci U S A. 2018 Jan 2;115(1):E82-E91. doi: 10.1073/pnas.1718441115. Epub 2017 Dec 18. Proc Natl Acad Sci U S A. 2018. PMID: 29255018 Free PMC article.
References
-
- Liu L, Fisher BE, Dowd KA, Astemborski J, Cox AL, Ray SC. 2010. Acceleration of hepatitis C virus envelope evolution in humans is consistent with progressive humoral immune selection during the transition from acute to chronic infection. J Virol 84:5067–5077. doi:10.1128/JVI.02265-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

