Methylthioadenosine (MTA) Regulates Liver Cells Proteome and Methylproteome: Implications in Liver Biology and Disease
- PMID: 26819315
- PMCID: PMC4858935
- DOI: 10.1074/mcp.M115.055772
Methylthioadenosine (MTA) Regulates Liver Cells Proteome and Methylproteome: Implications in Liver Biology and Disease
Abstract
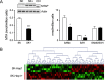
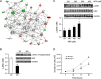
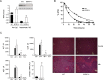
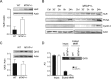
Methylthioadenosine phosphorylase (MTAP), a key enzyme in the adenine and methionine salvage pathways, catalyzes the hydrolysis of methylthioadenosine (MTA), a compound suggested to affect pivotal cellular processes in part through the regulation of protein methylation. MTAP is expressed in a wide range of cell types and tissues, and its deletion is common to cancer cells and in liver injury. The aim of this study was to investigate the proteome and methyl proteome alterations triggered by MTAP deficiency in liver cells to define novel regulatory mechanisms that may explain the pathogenic processes of liver diseases. iTRAQ analysis resulted in the identification of 216 differential proteins (p < 0.05) that suggest deregulation of cellular pathways as those mediated by ERK or NFκB. R-methyl proteome analysis led to the identification of 74 differentially methylated proteins between SK-Hep1 and SK-Hep1+ cells, including 116 new methylation sites. Restoring normal MTA levels in SK-Hep1+ cells parallels the specific methylation of 56 proteins, including KRT8, TGF, and CTF8A, which provides a novel regulatory mechanism of their activity with potential implications in carcinogenesis. Inhibition of RNA-binding proteins methylation is especially relevant upon accumulation of MTA. As an example, methylation of quaking protein in Arg(242) and Arg(256) in SK-Hep1+ cells may play a pivotal role in the regulation of its activity as indicated by the up-regulation of its target protein p27(kip1) The phenotype associated with a MTAP deficiency was further verified in the liver of MTAP± mice. Our data support that MTAP deficiency leads to MTA accumulation and deregulation of central cellular pathways, increasing proliferation and decreasing the susceptibility to chemotherapeutic drugs, which involves differential protein methylation. Data are available via ProteomeXchange with identifier PXD002957 (http://www.ebi.ac.uk/pride/archive/projects/PXD002957).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Down-regulation of methylthioadenosine phosphorylase (MTAP) induces progression of hepatocellular carcinoma via accumulation of 5'-deoxy-5'-methylthioadenosine (MTA).Am J Pathol. 2011 Mar;178(3):1145-52. doi: 10.1016/j.ajpath.2010.11.059. Epub 2011 Jan 28. Am J Pathol. 2011. PMID: 21356366 Free PMC article.
-
Extracellular 5'-methylthioadenosine inhibits intracellular symmetric dimethylarginine protein methylation of FUSE-binding proteins.J Biol Chem. 2022 Sep;298(9):102367. doi: 10.1016/j.jbc.2022.102367. Epub 2022 Aug 11. J Biol Chem. 2022. PMID: 35963436 Free PMC article.
-
Homozygous MTAP deletion in primary human glioblastoma is not associated with elevation of methylthioadenosine.Nat Commun. 2021 Jul 9;12(1):4228. doi: 10.1038/s41467-021-24240-3. Nat Commun. 2021. PMID: 34244484 Free PMC article.
-
Targeting tumors that lack methylthioadenosine phosphorylase (MTAP) activity: current strategies.Cancer Biol Ther. 2011 Apr 1;11(7):627-32. doi: 10.4161/cbt.11.7.14948. Epub 2011 Apr 1. Cancer Biol Ther. 2011. PMID: 21301207 Free PMC article. Review.
-
Methylthioadenosine.Int J Biochem Cell Biol. 2004 Nov;36(11):2125-30. doi: 10.1016/j.biocel.2003.11.016. Int J Biochem Cell Biol. 2004. PMID: 15313459 Review.
Cited by
-
Targeted Proteomics for Monitoring One-Carbon Metabolism in Liver Diseases.Metabolites. 2022 Aug 24;12(9):779. doi: 10.3390/metabo12090779. Metabolites. 2022. PMID: 36144184 Free PMC article. Review.
-
Excess S-adenosylmethionine inhibits methylation via catabolism to adenine.Commun Biol. 2022 Apr 5;5(1):313. doi: 10.1038/s42003-022-03280-5. Commun Biol. 2022. PMID: 35383287 Free PMC article.
-
Reduced symmetric dimethylation stabilizes vimentin and promotes metastasis in MTAP-deficient lung cancer.EMBO Rep. 2022 Aug 3;23(8):e54265. doi: 10.15252/embr.202154265. Epub 2022 Jun 29. EMBO Rep. 2022. PMID: 35766227 Free PMC article.
-
Solid phase microextraction chemical biopsy tool for monitoring of doxorubicin residue during in vivo lung chemo-perfusion.J Pharm Anal. 2021 Feb;11(1):37-47. doi: 10.1016/j.jpha.2020.08.011. Epub 2020 Sep 2. J Pharm Anal. 2021. PMID: 33717610 Free PMC article.
-
Highlights of the Biology and Disease-driven Human Proteome Project, 2015-2016.J Proteome Res. 2016 Nov 4;15(11):3979-3987. doi: 10.1021/acs.jproteome.6b00444. Epub 2016 Sep 20. J Proteome Res. 2016. PMID: 27573249 Free PMC article.
References
-
- Williams R. (2006) Global challenges in liver disease. Hepatology 44, 521–526 - PubMed
-
- Berasain C., Sampedro A., Mauleon I., Goni S., Latasa M. U., Matscheko N., Garcia-Bravo M., Unzu C., Corrales F. J., Enriquez de Salamanca R., Prieto J., Avila M. A., and Fontanellas A. (2009) Epidermal growth factor receptor ligands in murine models for erythropoietic protoporphyria: Potential novel players in the progression of liver injury. Cell. Mol. Biol. 55, 29–37 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

