PDIP46 (DNA polymerase δ interacting protein 46) is an activating factor for human DNA polymerase δ
- PMID: 26819372
- PMCID: PMC4868757
- DOI: 10.18632/oncotarget.7034
PDIP46 (DNA polymerase δ interacting protein 46) is an activating factor for human DNA polymerase δ
Abstract
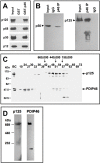
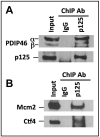
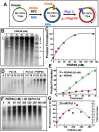
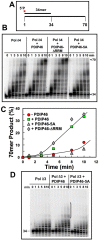
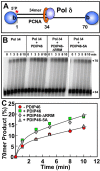
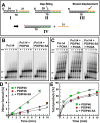
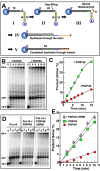
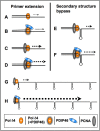
PDIP46 (SKAR, POLDIP3) was discovered through its interaction with the p50 subunit of human DNA polymerase δ (Pol δ). Its functions in DNA replication are unknown. PDIP46 associates with Pol δ in cell extracts both by immunochemical and protein separation methods, as well as by ChIP analyses. PDIP46 also interacts with PCNA via multiple copies of a novel PCNA binding motif, the APIMs (AlkB homologue-2 PCNA-Interacting Motif). Sites for both p50 and PCNA binding were mapped to the N-terminal region containing the APIMs. Functional assays for the effects of PDIP46 on Pol δ activity on singly primed ssM13 DNA templates revealed that it is a novel and potent activator of Pol δ. The effects of PDIP46 on Pol δ in primer extension, strand displacement and synthesis through simple hairpin structures reveal a mechanism where PDIP46 facilitates Pol δ4 synthesis through regions of secondary structure on complex templates. In addition, evidence was obtained that PDIP46 is also capable of exerting its effects by a direct interaction with Pol δ, independent of PCNA. Mutation of the Pol δ and PCNA binding region resulted in a loss of PDIP46 functions. These studies support the view that PDIP46 is a novel accessory protein for Pol δ that is involved in cellular DNA replication. This raises the possibility that altered expression of PDIP46 or its mutation may affect Pol δ functions in vivo, and thereby be a nexus for altered genomic stability.
Keywords: DNA polymerase δ; DNA replication; PDIP46; POLDIP3; SKAR.
Conflict of interest statement
None
Figures


Similar articles
-
Human enhancer of rudimentary is a molecular partner of PDIP46/SKAR, a protein interacting with DNA polymerase delta and S6K1 and regulating cell growth.FEBS J. 2006 Oct;273(20):4728-41. doi: 10.1111/j.1742-4658.2006.05477.x. Epub 2006 Sep 19. FEBS J. 2006. PMID: 16984396
-
Regulation and Modulation of Human DNA Polymerase δ Activity and Function.Genes (Basel). 2017 Jul 24;8(7):190. doi: 10.3390/genes8070190. Genes (Basel). 2017. PMID: 28737709 Free PMC article. Review.
-
A tumor necrosis factor alpha- and interleukin 6-inducible protein that interacts with the small subunit of DNA polymerase delta and proliferating cell nuclear antigen.Proc Natl Acad Sci U S A. 2001 Oct 9;98(21):11979-84. doi: 10.1073/pnas.221452098. Proc Natl Acad Sci U S A. 2001. PMID: 11593007 Free PMC article.
-
Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen.J Biol Chem. 2003 Mar 21;278(12):10041-7. doi: 10.1074/jbc.M208694200. Epub 2003 Jan 9. J Biol Chem. 2003. PMID: 12522211
-
Two forms of human DNA polymerase δ: Who does what and why?DNA Repair (Amst). 2019 Sep;81:102656. doi: 10.1016/j.dnarep.2019.102656. Epub 2019 Jul 8. DNA Repair (Amst). 2019. PMID: 31326365 Review.
Cited by
-
DNA polymerase delta interacting protein 3 facilitates the activation and maintenance of DNA damage checkpoint in response to replication stress.Animal Model Exp Med. 2022 Oct;5(5):461-469. doi: 10.1002/ame2.12274. Epub 2022 Sep 27. Animal Model Exp Med. 2022. PMID: 36168146 Free PMC article.
-
Identification of necroptosis-related diagnostic biomarkers in coronary heart disease.Heliyon. 2024 Apr 25;10(9):e30269. doi: 10.1016/j.heliyon.2024.e30269. eCollection 2024 May 15. Heliyon. 2024. PMID: 38726127 Free PMC article.
-
POLDIP3: At the Crossroad of RNA and DNA Metabolism.Genes (Basel). 2022 Oct 22;13(11):1921. doi: 10.3390/genes13111921. Genes (Basel). 2022. PMID: 36360158 Free PMC article. Review.
-
Loss of the p12 subunit of DNA polymerase delta leads to a defect in HR and sensitization to PARP inhibitors.DNA Repair (Amst). 2019 Jan;73:64-70. doi: 10.1016/j.dnarep.2018.11.003. Epub 2018 Nov 13. DNA Repair (Amst). 2019. PMID: 30470508 Free PMC article.
-
The p12 Subunit Choreographs the Regulation and Functions of Two Forms of DNA Polymerase δ in Mammalian Cells.Genes (Basel). 2025 Feb 3;16(2):188. doi: 10.3390/genes16020188. Genes (Basel). 2025. PMID: 40004517 Free PMC article. Review.
References
-
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. - PubMed
-
- Loeb LA, Monnat RJJ. DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. - PubMed
-
- Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804:1049–1063. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

