Efficient enzymatic synthesis and dual-colour fluorescent labelling of DNA probes using long chain azido-dUTP and BCN dyes
- PMID: 26819406
- PMCID: PMC4856977
- DOI: 10.1093/nar/gkw028
Efficient enzymatic synthesis and dual-colour fluorescent labelling of DNA probes using long chain azido-dUTP and BCN dyes
Abstract
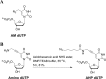
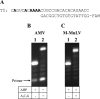
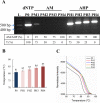
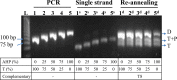
A sterically undemanding azide analogue of dTTP (AHP dUTP) with an alkyl chain and ethynyl attachment to the nucleobase was designed and incorporated into DNA by primer extension, reverse transcription and polymerase chain reaction (PCR). An azide-modified 523 bp PCR amplicon with all 335 thymidines replaced by AHP dU was shown to be a perfect copy of the template from which it was amplified. Replacement of thymidine with AHP dU increases duplex stability, accounting in part for the high incorporation efficiency of the azide-modified triphosphate. Single-stranded azide-labelled DNA was conveniently prepared from PCR products by λ-exonuclease digestion and streptavidin magnetic bead isolation. Efficient fluorescent labelling of single and double-stranded DNA was carried out using dyes functionalized with bicyclo[6.1.0]non-4-yne (BCN) via the strain-promoted alkyne-azide cycloaddition (SPAAC) reaction. This revealed that the degree of labelling must be carefully controlled to achieve optimum fluorescence and avoid fluorescence quenching. Dual-coloured probes were obtained in a single tube fluorescent labelling reaction; and varying the ratios of the two dyes provides a simple method to prepare DNA probes with unique fluorescent signatures. AHP dUTP is a versatile clickable nucleotide with potentially wide applications in biology and nanotechnology including single molecule studies and synthesis of modified aptamer libraries via SELEX.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures








Similar articles
-
Azide and trans-cyclooctene dUTPs: incorporation into DNA probes and fluorescent click-labelling.Analyst. 2015 Apr 21;140(8):2671-8. doi: 10.1039/c5an00158g. Analyst. 2015. PMID: 25734317
-
Enzymatic incorporation and fluorescent labelling of cyclooctyne-modified deoxyuridine triphosphates in DNA.Bioorg Med Chem. 2014 Aug 15;22(16):4384-90. doi: 10.1016/j.bmc.2014.05.050. Epub 2014 Jun 5. Bioorg Med Chem. 2014. PMID: 24953951
-
Fluorescent labelling of in situ hybridisation probes through the copper-catalysed azide-alkyne cycloaddition reaction.Chromosome Res. 2016 Sep;24(3):299-307. doi: 10.1007/s10577-016-9522-z. Epub 2016 Apr 19. Chromosome Res. 2016. PMID: 27095480
-
Multifluorinated Aryl Azides for the Development of Improved H2 S Probes, and Fast Strain-promoted Azide-Alkyne Cycloaddition and Staudinger Reactions.Chem Asian J. 2020 May 4;15(9):1420-1429. doi: 10.1002/asia.202000005. Epub 2020 Mar 23. Chem Asian J. 2020. PMID: 32144862 Review.
-
Oligothiophenes as fluorescent markers for biological applications.Molecules. 2012 Jan 18;17(1):910-33. doi: 10.3390/molecules17010910. Molecules. 2012. PMID: 22258339 Free PMC article. Review.
Cited by
-
Nucleotide-Bearing Benzylidene-Tetrahydroxanthylium Near-IR Fluorophore for Sensing DNA Replication, Secondary Structures and Interactions.Chemistry. 2020 Sep 16;26(52):11950-11954. doi: 10.1002/chem.202003192. Epub 2020 Aug 17. Chemistry. 2020. PMID: 32633433 Free PMC article.
-
Expanding the chemical functionality of DNA nanomaterials generated by rolling circle amplification.Nucleic Acids Res. 2021 Sep 20;49(16):9042-9052. doi: 10.1093/nar/gkab720. Nucleic Acids Res. 2021. PMID: 34403467 Free PMC article.
-
1,3-Diketone-Modified Nucleotides and DNA for Cross-Linking with Arginine-Containing Peptides and Proteins.Angew Chem Int Ed Engl. 2021 Aug 2;60(32):17383-17387. doi: 10.1002/anie.202105126. Epub 2021 Jul 2. Angew Chem Int Ed Engl. 2021. PMID: 34107150 Free PMC article.
-
2-Substituted dATP Derivatives as Building Blocks for Polymerase-Catalyzed Synthesis of DNA Modified in the Minor Groove.Angew Chem Int Ed Engl. 2016 Dec 19;55(51):15856-15859. doi: 10.1002/anie.201609007. Epub 2016 Nov 23. Angew Chem Int Ed Engl. 2016. PMID: 27879047 Free PMC article.
-
Stable immobilization of aldehyde ketone reductase mutants containing nonstandard amino acids on an epoxy resin via strain-promoted alkyne-azide cycloaddition.RSC Adv. 2020 Jan 14;10(5):2624-2633. doi: 10.1039/c9ra09067c. eCollection 2020 Jan 14. RSC Adv. 2020. PMID: 35496112 Free PMC article.
References
-
- Wagner M., Horn M., Daims H. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr. Opin. Microbiol. 2003;6:302–309. - PubMed
-
- Ranasinghe R.T., Brown T. Fluorescence based strategies for genetic analysis. Chem. Commun. 2005:5487–5502. - PubMed
-
- Bailey S.M., Goodwin E.H., Cornforth M.N. Strand-specific fluorescence in situ hybridization: the CO-FISH family. Cytogenet. Genome Res. 2004;107:14–17. - PubMed
-
- Spear R.N., Li S.X., Nordheim E.V., Andrews J.H. Quantitative imaging and statistical analysis of fluorescence in situ hybridization (FISH) of Aureobasidium pullulans. J. Microbiol. Methods. 1999;35:101–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

