Symbiotic Bacteria in Gills and Guts of Chinese Mitten Crab (Eriocheir sinensis) Differ from the Free-Living Bacteria in Water
- PMID: 26820139
- PMCID: PMC4731060
- DOI: 10.1371/journal.pone.0148135
Symbiotic Bacteria in Gills and Guts of Chinese Mitten Crab (Eriocheir sinensis) Differ from the Free-Living Bacteria in Water
Abstract
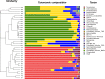
Aquatic animals have a close relationship with water, but differences in their symbiotic bacteria and the bacterial composition in water remains unclear. Wild or domestic Chinese mitten crabs (Eriocheir sinensis) and the water in which they live were collected from four sampling sites in Jiangsu and Shanghai, China. Bacterial composition in water, gills or guts of E. sinensis, were compared by high-throughput sequencing using 16S rRNA genes. Analysis of >660,000 sequences indicated that bacterial diversity was higher in water than in gills or guts. Tenericutes and Proteobacteria were dominant phyla in guts, while Actinobacteria, Proteobacteria and Bacteroidetes were dominant in gills and water. Non-metric multidimensional scaling analysis indicated that microbiota from gills, guts or water clearly separated into three groups, suggesting that crabs harbor a more specific microbial community than the water in which they live. The dominant OTUs in crab gut were related to Mycoplasmataceae, which were low in abundance in gills, showing that, like mammals, crabs have body-site specific microbiota. OTUs related to Ilumatobacter and Albimonas, which are commonly present in sediment and seawater, were dominant in gills but almost absent from the sampled water. Considering E. sinensis are bottom-dwelling crustacean and they mate in saline water or seawater, behavior and life cycle of crabs may play an important role in shaping the symbiotic bacterial pattern. This study revealed the relationship between the symbiotic bacteria of Chinese mitten crab and their habitat, affording information on the assembly factors of commensal bacteria in aquatic animals.
Conflict of interest statement
Figures


Similar articles
-
Phylogenetic analysis of intestinal bacteria in the Chinese mitten crab (Eriocheir sinensis).J Appl Microbiol. 2007 Sep;103(3):675-82. doi: 10.1111/j.1365-2672.2007.03295.x. J Appl Microbiol. 2007. PMID: 17714401
-
Seasonal dynamics of intestinal microbiota in juvenile Chinese mitten crab (Eriocheir sinensis) in the Yangtze Estuary.Front Cell Infect Microbiol. 2024 Jul 4;14:1436547. doi: 10.3389/fcimb.2024.1436547. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39027136 Free PMC article.
-
Changes in the gut microbiome of the Chinese mitten crab (Eriocheir sinensis) in response to White spot syndrome virus (WSSV) infection.J Fish Dis. 2017 Nov;40(11):1561-1571. doi: 10.1111/jfd.12624. Epub 2017 Apr 21. J Fish Dis. 2017. PMID: 28429823
-
Exploring prokaryotic diversity in the genomic era.Genome Biol. 2002;3(2):REVIEWS0003. doi: 10.1186/gb-2002-3-2-reviews0003. Epub 2002 Jan 29. Genome Biol. 2002. PMID: 11864374 Free PMC article. Review.
-
Current Disease Threats for Cultivated Crab Eriocheir sinensis in China.Transbound Emerg Dis. 2023 Apr 3;2023:3305963. doi: 10.1155/2023/3305963. eCollection 2023. Transbound Emerg Dis. 2023. PMID: 40303730 Free PMC article. Review.
Cited by
-
Comparative analysis of the intestinal bacterial community and expression of gut immunity genes in the Chinese Mitten Crab (Eriocheir sinensis).AMB Express. 2018 Dec 13;8(1):192. doi: 10.1186/s13568-018-0722-0. AMB Express. 2018. PMID: 30547243 Free PMC article.
-
The role of gut microbial community and metabolomic shifts in adaptive resistance of Atlantic killifish (Fundulus heteroclitus) to polycyclic aromatic hydrocarbons.Sci Total Environ. 2021 Jul 1;776:145955. doi: 10.1016/j.scitotenv.2021.145955. Epub 2021 Feb 19. Sci Total Environ. 2021. PMID: 33647645 Free PMC article.
-
Microbiome of the Black-Lipped Pearl Oyster Pinctada margaritifera, a Multi-Tissue Description With Functional Profiling.Front Microbiol. 2019 Jul 5;10:1548. doi: 10.3389/fmicb.2019.01548. eCollection 2019. Front Microbiol. 2019. PMID: 31333634 Free PMC article.
-
The effect of dietary supplementation with Clostridium butyricum on the growth performance, immunity, intestinal microbiota and disease resistance of tilapia (Oreochromis niloticus).PLoS One. 2019 Dec 9;14(12):e0223428. doi: 10.1371/journal.pone.0223428. eCollection 2019. PLoS One. 2019. PMID: 31815958 Free PMC article.
-
Change in the intestinal bacterial community structure associated with environmental microorganisms during the growth of Eriocheir sinensis.Microbiologyopen. 2019 May;8(5):e00727. doi: 10.1002/mbo3.727. Epub 2018 Oct 11. Microbiologyopen. 2019. PMID: 30311433 Free PMC article.
References
-
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

