HPV Population Profiling in Healthy Men by Next-Generation Deep Sequencing Coupled with HPV-QUEST
- PMID: 26821041
- PMCID: PMC4776183
- DOI: 10.3390/v8020028
HPV Population Profiling in Healthy Men by Next-Generation Deep Sequencing Coupled with HPV-QUEST
Abstract
Multiple-type human papillomaviruses (HPV) infection presents a greater risk for persistence in asymptomatic individuals and may accelerate cancer development. To extend the scope of HPV types defined by probe-based assays, multiplexing deep sequencing of HPV L1, coupled with an HPV-QUEST genotyping server and a bioinformatic pipeline, was established and applied to survey the diversity of HPV genotypes among a subset of healthy men from the HPV in Men (HIM) Multinational Study. Twenty-one HPV genotypes (12 high-risk and 9 low-risk) were detected in the genital area from 18 asymptomatic individuals. A single HPV type, either HPV16, HPV6b or HPV83, was detected in 7 individuals, while coinfection by 2 to 5 high-risk and/or low-risk genotypes was identified in the other 11 participants. In two individuals studied for over one year, HPV16 persisted, while fluctuations of coinfecting genotypes occurred. HPV L1 regions were generally identical between query and reference sequences, although nonsynonymous and synonymous nucleotide polymorphisms of HPV16, 18, 31, 35h, 59, 70, 73, cand85, 6b, 62, 81, 83, cand89 or JEB2 L1 genotypes, mostly unidentified by linear array, were evident. Deep sequencing coupled with HPV-QUEST provides efficient and unambiguous classification of HPV genotypes in multiple-type HPV infection in host ecosystems.
Keywords: HPV; HPV-QUEST; Linear Array; Papillomavirus Episteme; asymptomatic men; deep sequencing; multiple-type HPV infection.
Figures
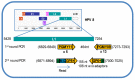
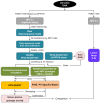

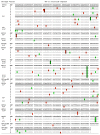

Similar articles
-
Human papillomavirus genotyping by Linear Array and Next-Generation Sequencing in cervical samples from Western Mexico.Virol J. 2015 Oct 6;12:161. doi: 10.1186/s12985-015-0391-4. Virol J. 2015. PMID: 26444975 Free PMC article.
-
Cross-hybridization between HPV genotypes in the Linear Array Genotyping Test confirmed by Next-Generation Sequencing.Diagn Pathol. 2019 Apr 22;14(1):31. doi: 10.1186/s13000-019-0808-2. Diagn Pathol. 2019. PMID: 31010421 Free PMC article.
-
Accurate human papillomavirus genotyping by 454 pyrosequencing.Clin Microbiol Infect. 2013 Oct;19(10):E428-34. doi: 10.1111/1469-0691.12219. Epub 2013 Apr 10. Clin Microbiol Infect. 2013. PMID: 23573945
-
Prevalence and genotype distribution of cervical human papillomavirus (HPV) among women in urban Tianjin, China.J Med Virol. 2015 Nov;87(11):1966-72. doi: 10.1002/jmv.24248. Epub 2015 Jun 12. J Med Virol. 2015. PMID: 26073652
-
Utility of high-throughput DNA sequencing in the study of the human papillomaviruses.Virus Genes. 2018 Feb;54(1):17-24. doi: 10.1007/s11262-017-1530-3. Epub 2017 Dec 27. Virus Genes. 2018. PMID: 29282656 Review.
Cited by
-
Universal human papillomavirus typing by whole genome sequencing following target enrichment: evaluation of assay reproducibility and limit of detection.BMC Genomics. 2019 Mar 20;20(1):231. doi: 10.1186/s12864-019-5598-0. BMC Genomics. 2019. PMID: 30894118 Free PMC article.
-
Universal Human Papillomavirus Typing Assay: Whole-Genome Sequencing following Target Enrichment.J Clin Microbiol. 2017 Mar;55(3):811-823. doi: 10.1128/JCM.02132-16. Epub 2016 Dec 14. J Clin Microbiol. 2017. PMID: 27974548 Free PMC article.
-
Broad-Spectrum Detection of HPV in Male Genital Samples Using Target-Enriched Whole-Genome Sequencing.Viruses. 2023 Sep 21;15(9):1967. doi: 10.3390/v15091967. Viruses. 2023. PMID: 37766373 Free PMC article.
-
HPV infection in semen: results from a new molecular approach.Epidemiol Infect. 2019 Jan;147:e177. doi: 10.1017/S0950268819000621. Epidemiol Infect. 2019. PMID: 31063107 Free PMC article.
-
Identification by high-throughput sequencing of HPV variants and quasispecies that are untypeable by linear reverse blotting assay in cervical specimens.Papillomavirus Res. 2019 Dec;8:100169. doi: 10.1016/j.pvr.2019.100169. Epub 2019 Jul 5. Papillomavirus Res. 2019. PMID: 31283993 Free PMC article.
References
-
- Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

