The contribution of Alu exons to the human proteome
- PMID: 26821878
- PMCID: PMC4731929
- DOI: 10.1186/s13059-016-0876-5
The contribution of Alu exons to the human proteome
Abstract
Background: Alu elements are major contributors to lineage-specific new exons in primate and human genomes. Recent studies indicate that some Alu exons have high transcript inclusion levels or tissue-specific splicing profiles, and may play important regulatory roles in modulating mRNA degradation or translational efficiency. However, the contribution of Alu exons to the human proteome remains unclear and controversial. The prevailing view is that exons derived from young repetitive elements, such as Alu elements, are restricted to regulatory functions and have not had adequate evolutionary time to be incorporated into stable, functional proteins.
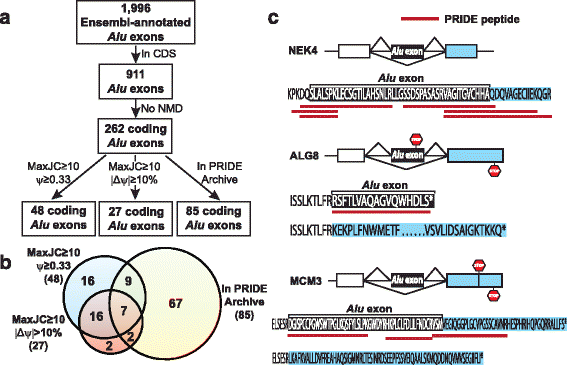
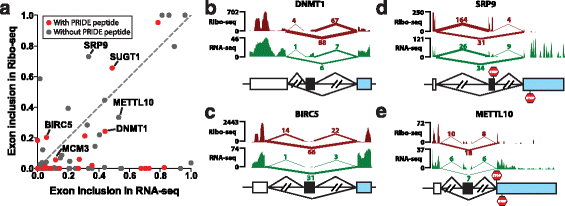
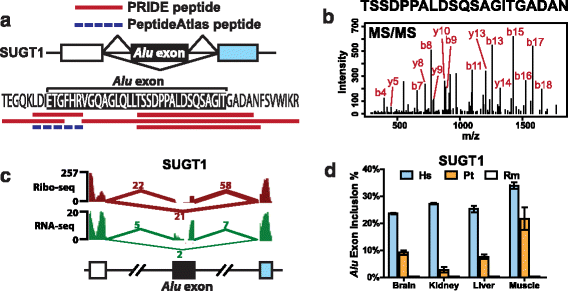
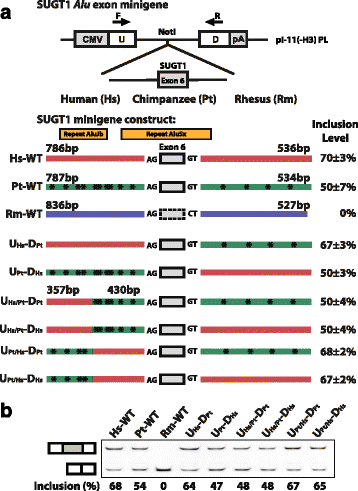
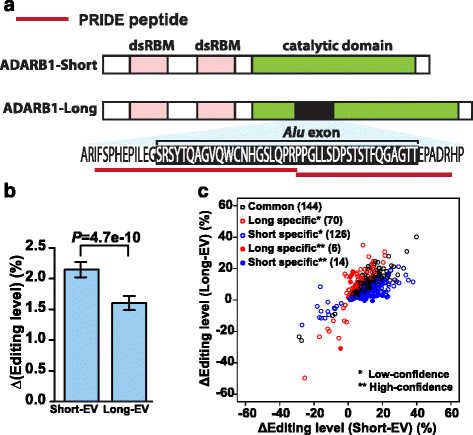
Results: We adopt a proteotranscriptomics approach to systematically assess the contribution of Alu exons to the human proteome. Using RNA sequencing, ribosome profiling, and proteomics data from human tissues and cell lines, we provide evidence for the translational activities of Alu exons and the presence of Alu exon derived peptides in human proteins. These Alu exon peptides represent species-specific protein differences between primates and other mammals, and in certain instances between humans and closely related primates. In the case of the RNA editing enzyme ADARB1, which contains an Alu exon peptide in its catalytic domain, RNA sequencing analyses of A-to-I editing demonstrate that both the Alu exon skipping and inclusion isoforms encode active enzymes. The Alu exon derived peptide may fine tune the overall editing activity and, in limited cases, the site selectivity of ADARB1 protein products.
Conclusions: Our data indicate that Alu elements have contributed to the acquisition of novel protein sequences during primate and human evolution.
Figures
Similar articles
-
RNA-editing-mediated exon evolution.Genome Biol. 2007;8(2):R29. doi: 10.1186/gb-2007-8-2-r29. Genome Biol. 2007. PMID: 17326827 Free PMC article.
-
DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome.Nature. 2017 Apr 6;544(7648):115-119. doi: 10.1038/nature21715. Epub 2017 Mar 29. Nature. 2017. PMID: 28355180
-
Diverse splicing patterns of exonized Alu elements in human tissues.PLoS Genet. 2008 Oct 17;4(10):e1000225. doi: 10.1371/journal.pgen.1000225. PLoS Genet. 2008. PMID: 18841251 Free PMC article.
-
ALU A-to-I RNA Editing: Millions of Sites and Many Open Questions.Methods Mol Biol. 2021;2181:149-162. doi: 10.1007/978-1-0716-0787-9_9. Methods Mol Biol. 2021. PMID: 32729079 Review.
-
Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome.EMBO Rep. 2005 Sep;6(9):831-5. doi: 10.1038/sj.embor.7400507. EMBO Rep. 2005. PMID: 16138094 Free PMC article. Review.
Cited by
-
Retrotransposon-induced mosaicism in the neural genome.Open Biol. 2018 Jul;8(7):180074. doi: 10.1098/rsob.180074. Open Biol. 2018. PMID: 30021882 Free PMC article. Review.
-
Alu RNA and their roles in human disease states.RNA Biol. 2021 Nov 12;18(sup2):574-585. doi: 10.1080/15476286.2021.1989201. Epub 2021 Oct 21. RNA Biol. 2021. PMID: 34672903 Free PMC article.
-
Warning SINEs: Alu elements, evolution of the human brain, and the spectrum of neurological disease.Chromosome Res. 2018 Mar;26(1-2):93-111. doi: 10.1007/s10577-018-9573-4. Epub 2018 Feb 19. Chromosome Res. 2018. PMID: 29460123 Free PMC article. Review.
-
Transcriptome protection by the expanded family of hnRNPs.RNA Biol. 2019 Feb;16(2):155-159. doi: 10.1080/15476286.2018.1564617. Epub 2019 Jan 6. RNA Biol. 2019. PMID: 30596342 Free PMC article. Review.
-
Few SINEs of life: Alu elements have little evidence for biological relevance despite elevated translation.NAR Genom Bioinform. 2020 Mar;2(1):lqz023. doi: 10.1093/nargab/lqz023. Epub 2019 Dec 19. NAR Genom Bioinform. 2020. PMID: 31886458 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

