A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease
- PMID: 26822079
- PMCID: PMC4740116
- DOI: 10.1038/ncomms10311
A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease
Abstract
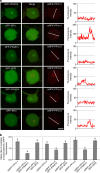
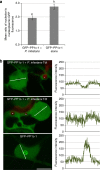
Plant pathogens deliver effectors to alter host processes. Knowledge of how effectors target and manipulate host proteins is critical to understand crop disease. Here, we show that in planta expression of the RXLR effector Pi04314 enhances leaf colonization by Phytophthora infestans via activity in the host nucleus and attenuates induction of jasmonic and salicylic acid-responsive genes. Pi04314 interacts with three host protein phosphatase 1 catalytic (PP1c) isoforms, causing their re-localization from the nucleolus to the nucleoplasm. Re-localization of PP1c-1 also occurs during infection and is dependent on an R/KVxF motif in the effector. Silencing the PP1c isoforms or overexpression of a phosphatase-dead PP1c-1 mutant attenuates infection, demonstrating that host PP1c activity is required for disease. Moreover, expression of PP1c-1mut abolishes enhanced leaf colonization mediated by in planta Pi04314 expression. We argue that PP1c isoforms are susceptibility factors forming holoenzymes with Pi04314 to promote late blight disease.
Figures



References
-
- Dou D. & Zhou J.-M. Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495 (2012). - PubMed
-
- Jones J. D. & Dangl J. L. The plant immune system. Nature 444, 323–329 (2006). - PubMed
-
- Deslandes L. & Rivas S. Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17, 644–655 (2012). - PubMed
-
- Win J. et al.. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77, 235–247 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

