MTD: a mammalian transcriptomic database to explore gene expression and regulation
- PMID: 26822098
- PMCID: PMC5221423
- DOI: 10.1093/bib/bbv117
MTD: a mammalian transcriptomic database to explore gene expression and regulation
Abstract
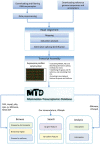
A systematic transcriptome survey is essential for the characterization and comprehension of the molecular basis underlying phenotypic variations. Recently developed RNA-seq methodology has facilitated efficient data acquisition and information mining of transcriptomes in multiple tissues/cell lines. Current mammalian transcriptomic databases are either tissue-specific or species-specific, and they lack in-depth comparative features across tissues and species. Here, we present a mammalian transcriptomic database (MTD) that is focused on mammalian transcriptomes, and the current version contains data from humans, mice, rats and pigs. Regarding the core features, the MTD browses genes based on their neighboring genomic coordinates or joint KEGG pathway and provides expression information on exons, transcripts and genes by integrating them into a genome browser. We developed a novel nomenclature for each transcript that considers its genomic position and transcriptional features. The MTD allows a flexible search of genes or isoforms with user-defined transcriptional characteristics and provides both table-based descriptions and associated visualizations. To elucidate the dynamics of gene expression regulation, the MTD also enables comparative transcriptomic analysis in both intraspecies and interspecies manner. The MTD thus constitutes a valuable resource for transcriptomic and evolutionary studies. The MTD is freely accessible at http://mtd.cbi.ac.cn.
Keywords: RNA-seq; gene expression and regulation; mammalian transcriptomic database.
© The Author 2016. Published by Oxford University Press.
Figures


Similar articles
-
Express: A database of transcriptome profiles encompassing known and novel transcripts across multiple development stages in eye tissues.Exp Eye Res. 2018 Mar;168:57-68. doi: 10.1016/j.exer.2018.01.009. Epub 2018 Jan 11. Exp Eye Res. 2018. PMID: 29337142 Free PMC article.
-
GATExplorer: genomic and transcriptomic explorer; mapping expression probes to gene loci, transcripts, exons and ncRNAs.BMC Bioinformatics. 2010 Apr 29;11:221. doi: 10.1186/1471-2105-11-221. BMC Bioinformatics. 2010. PMID: 20429936 Free PMC article.
-
Nicotiana attenuata Data Hub (NaDH): an integrative platform for exploring genomic, transcriptomic and metabolomic data in wild tobacco.BMC Genomics. 2017 Jan 13;18(1):79. doi: 10.1186/s12864-016-3465-9. BMC Genomics. 2017. PMID: 28086860 Free PMC article.
-
Characterizing and annotating the genome using RNA-seq data.Sci China Life Sci. 2017 Feb;60(2):116-125. doi: 10.1007/s11427-015-0349-4. Epub 2016 Jun 13. Sci China Life Sci. 2017. PMID: 27294835 Review.
-
A pulmonologist's guide to perform and analyse cross-species single lung cell transcriptomics.Eur Respir Rev. 2022 Jul 27;31(165):220056. doi: 10.1183/16000617.0056-2022. Print 2022 Sep 30. Eur Respir Rev. 2022. PMID: 35896273 Free PMC article. Review.
Cited by
-
The BIG Data Center: from deposition to integration to translation.Nucleic Acids Res. 2017 Jan 4;45(D1):D18-D24. doi: 10.1093/nar/gkw1060. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899658 Free PMC article.
-
Systematic integrated analysis of genetic and epigenetic variation in diabetic kidney disease.Proc Natl Acad Sci U S A. 2020 Nov 17;117(46):29013-29024. doi: 10.1073/pnas.2005905117. Epub 2020 Nov 3. Proc Natl Acad Sci U S A. 2020. PMID: 33144501 Free PMC article.
-
In silico design of context-responsive mammalian promoters with user-defined functionality.Nucleic Acids Res. 2017 Oct 13;45(18):10906-10919. doi: 10.1093/nar/gkx768. Nucleic Acids Res. 2017. PMID: 28977454 Free PMC article.
-
ANGIOGENES: knowledge database for protein-coding and noncoding RNA genes in endothelial cells.Sci Rep. 2016 Sep 1;6:32475. doi: 10.1038/srep32475. Sci Rep. 2016. PMID: 27582018 Free PMC article.
-
Processing and Analysis of RNA-seq Data from Public Resources.Methods Mol Biol. 2021;2243:81-94. doi: 10.1007/978-1-0716-1103-6_4. Methods Mol Biol. 2021. PMID: 33606253 Review.
References
-
- Adams J. Transcriptome: connecting the genome to gene function. Nat Educ 2008;1:195.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

