Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors
- PMID: 26822237
- PMCID: PMC5471125
- DOI: 10.1001/jamaoncol.2015.5699
Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors
Abstract
Importance: Whole-exome sequencing (WES) has the potential to reveal tumor and germline mutations of clinical relevance, but the diagnostic yield for pediatric patients with solid tumors is unknown.
Objective: To characterize the diagnostic yield of combined tumor and germline WES for children with solid tumors.
Design: Unselected children with newly diagnosed and previously untreated central nervous system (CNS) and non-CNS solid tumors were prospectively enrolled in the BASIC3 study at a large academic children's hospital during a 23-month period from August 2012 through June 2014. Blood and tumor samples underwent WES in a certified clinical laboratory with genetic results categorized on the basis of perceived clinical relevance and entered in the electronic health record.
Main outcomes and measures: Clinical categorization of somatic mutations; frequencies of deleterious germline mutations related to patient phenotype and incidental medically-actionable mutations.
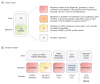
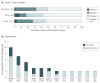
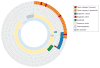
Results: Of the first 150 participants (80 boys and 70 girls, mean age, 7.4 years), tumor samples adequate for WES were available from 121 patients (81%). Somatic mutations of established clinical utility (category I) were reported in 4 (3%) of 121 patients, with mutations of potential utility (category II) detected in an additional 29 (24%) of 121 patients. CTNNB1 was the gene most frequently mutated, with recurrent mutations in KIT, TSC2, and MAPK pathway genes (BRAF, KRAS, and NRAS) also identified. Mutations in consensus cancer genes (category III) were found in an additional 24 (20%) of 121 tumors. Fewer than half of somatic mutations identified were in genes known to be recurrently mutated in the tumor type tested. Diagnostic germline findings related to patient phenotype were discovered in 15 (10%) of 150 cases: 13 pathogenic or likely pathogenic dominant mutations in adult and pediatric cancer susceptibility genes (including 2 each in TP53, VHL, and BRCA1), 1 recessive liver disorder with hepatocellular carcinoma (TJP2), and 1 renal diagnosis (CLCN5). Incidental findings were reported in 8 (5%) of 150 patients. Most patients harbored germline uncertain variants in cancer genes (98%), pharmacogenetic variants (89%), and recessive carrier mutations (85%).
Conclusions and relevance: Tumor and germline WES revealed mutations in a broad spectrum of genes previously implicated in both adult and pediatric cancers. Combined reporting of tumor and germline WES identified diagnostic and/or potentially actionable findings in nearly 40% of newly diagnosed pediatric patients with solid tumors.
Conflict of interest statement
Figures
References
-
- Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31(15):1806–1814. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

