A multi-substrate approach for functional metagenomics-based screening for (hemi)cellulases in two wheat straw-degrading microbial consortia unveils novel thermoalkaliphilic enzymes
- PMID: 26822785
- PMCID: PMC4730625
- DOI: 10.1186/s12864-016-2404-0
A multi-substrate approach for functional metagenomics-based screening for (hemi)cellulases in two wheat straw-degrading microbial consortia unveils novel thermoalkaliphilic enzymes
Abstract
Background: Functional metagenomics is a promising strategy for the exploration of the biocatalytic potential of microbiomes in order to uncover novel enzymes for industrial processes (e.g. biorefining or bleaching pulp). Most current methodologies used to screen for enzymes involved in plant biomass degradation are based on the use of single substrates. Moreover, highly diverse environments are used as metagenomic sources. However, such methods suffer from low hit rates of positive clones and hence the discovery of novel enzymatic activities from metagenomes has been hampered.
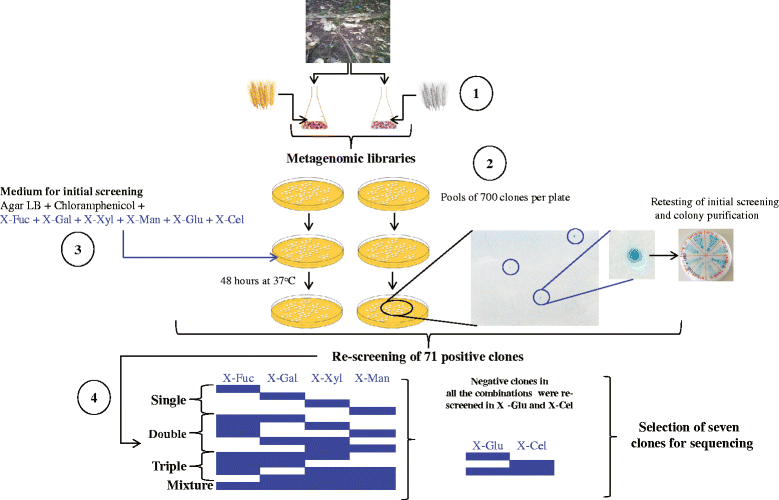
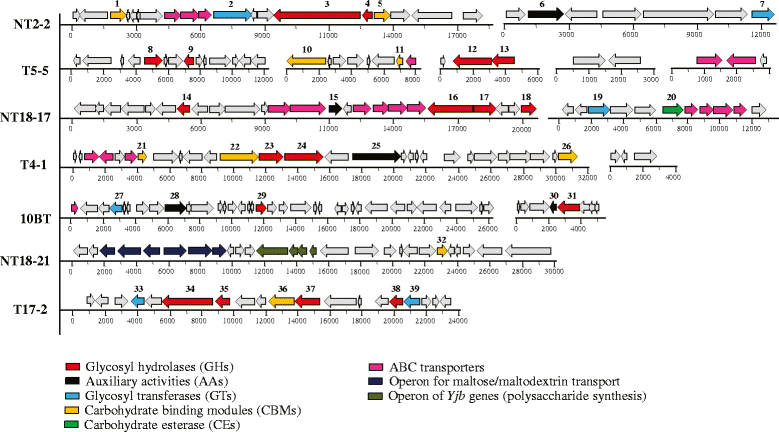
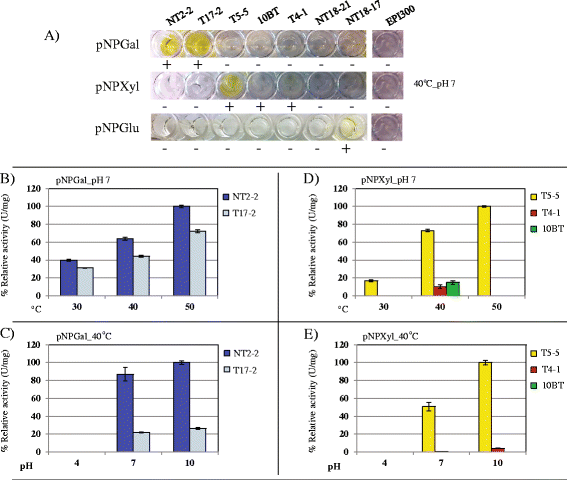
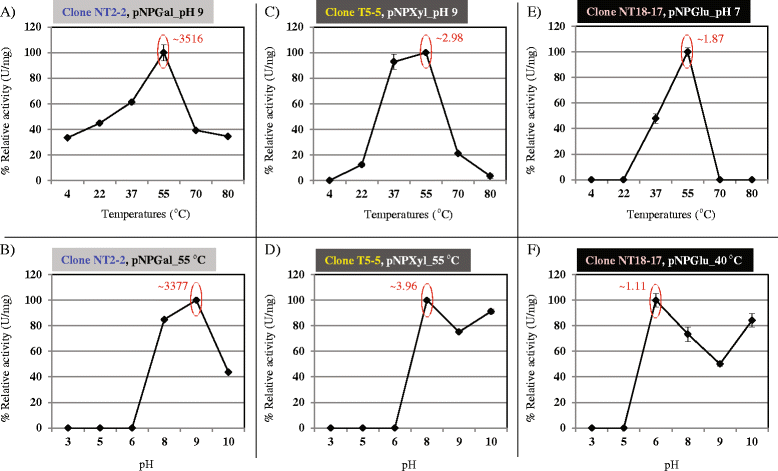
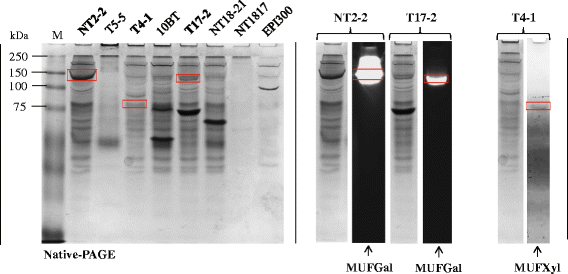
Results: Here, we constructed fosmid libraries from two wheat straw-degrading microbial consortia, denoted RWS (bred on untreated wheat straw) and TWS (bred on heat-treated wheat straw). Approximately 22,000 clones from each library were screened for (hemi)cellulose-degrading enzymes using a multi-chromogenic substrate approach. The screens yielded 71 positive clones for both libraries, giving hit rates of 1:440 and 1:1,047 for RWS and TWS, respectively. Seven clones (NT2-2, T5-5, NT18-17, T4-1, 10BT, NT18-21 and T17-2) were selected for sequence analyses. Their inserts revealed the presence of 18 genes encoding enzymes belonging to twelve different glycosyl hydrolase families (GH2, GH3, GH13, GH17, GH20, GH27, GH32, GH39, GH53, GH58, GH65 and GH109). These encompassed several carbohydrate-active gene clusters traceable mainly to Klebsiella related species. Detailed functional analyses showed that clone NT2-2 (containing a beta-galactosidase of ~116 kDa) had highest enzymatic activity at 55 °C and pH 9.0. Additionally, clone T5-5 (containing a beta-xylosidase of ~86 kDa) showed > 90% of enzymatic activity at 55 °C and pH 10.0.
Conclusions: This study employed a high-throughput method for rapid screening of fosmid metagenomic libraries for (hemi)cellulose-degrading enzymes. The approach, consisting of screens on multi-substrates coupled to further analyses, revealed high hit rates, as compared with recent other studies. Two clones, 10BT and T4-1, required the presence of multiple substrates for detectable activity, indicating a new avenue in library activity screening. Finally, clones NT2-2, T5-5 and NT18-17 were found to encode putative novel thermo-alkaline enzymes, which could represent a starting point for further biotechnological applications.
Figures
Similar articles
-
Unveiling the metabolic potential of two soil-derived microbial consortia selected on wheat straw.Sci Rep. 2015 Sep 7;5:13845. doi: 10.1038/srep13845. Sci Rep. 2015. PMID: 26343383 Free PMC article.
-
Applying functional metagenomics to search for novel lignocellulosic enzymes in a microbial consortium derived from a thermophilic composting phase of sugarcane bagasse and cow manure.Antonie Van Leeuwenhoek. 2016 Sep;109(9):1217-33. doi: 10.1007/s10482-016-0723-4. Epub 2016 Jun 27. Antonie Van Leeuwenhoek. 2016. PMID: 27350392
-
Identification and characterization of a cellulase-encoding gene from the buffalo rumen metagenomic library.Biosci Biotechnol Biochem. 2012;76(6):1075-84. doi: 10.1271/bbb.110786. Epub 2012 Jun 7. Biosci Biotechnol Biochem. 2012. PMID: 22790926
-
Bioprospecting of functional cellulases from metagenome for second generation biofuel production: a review.Crit Rev Microbiol. 2018 Mar;44(2):244-257. doi: 10.1080/1040841X.2017.1337713. Epub 2017 Jun 13. Crit Rev Microbiol. 2018. PMID: 28609211 Review.
-
Metagenomics: Probing pollutant fate in natural and engineered ecosystems.Biotechnol Adv. 2016 Dec;34(8):1413-1426. doi: 10.1016/j.biotechadv.2016.10.006. Epub 2016 Nov 5. Biotechnol Adv. 2016. PMID: 27825829 Review.
Cited by
-
Substrate Shift Reveals Roles for Members of Bacterial Consortia in Degradation of Plant Cell Wall Polymers.Front Microbiol. 2018 Mar 1;9:364. doi: 10.3389/fmicb.2018.00364. eCollection 2018. Front Microbiol. 2018. PMID: 29545786 Free PMC article.
-
Temporal Expression Dynamics of Plant Biomass-Degrading Enzymes by a Synthetic Bacterial Consortium Growing on Sugarcane Bagasse.Front Microbiol. 2018 Feb 26;9:299. doi: 10.3389/fmicb.2018.00299. eCollection 2018. Front Microbiol. 2018. PMID: 29535687 Free PMC article.
-
Multifunctional cellulases are potent, versatile tools for a renewable bioeconomy.Curr Opin Biotechnol. 2021 Feb;67:141-148. doi: 10.1016/j.copbio.2020.12.020. Epub 2021 Feb 4. Curr Opin Biotechnol. 2021. PMID: 33550093 Free PMC article. Review.
-
Study on the effect of microbial agent M44 on straw decay promoting in cold soils.Sci Rep. 2025 Jul 2;15(1):22539. doi: 10.1038/s41598-025-05817-0. Sci Rep. 2025. PMID: 40593055 Free PMC article.
-
Functional metagenomics identifies an exosialidase with an inverting catalytic mechanism that defines a new glycoside hydrolase family (GH156).J Biol Chem. 2018 Nov 23;293(47):18138-18150. doi: 10.1074/jbc.RA118.003302. Epub 2018 Sep 24. J Biol Chem. 2018. PMID: 30249617 Free PMC article.
References
-
- Limayem A, Ricke SC. Lignocellulosic biomass for bioethanol production: Current perspectives, potential issues and future prospects. Prog Energy Combust Sci. 2012;38:449–67. doi: 10.1016/j.pecs.2012.03.002. - DOI
-
- Himmel ME, Xu Q, Luo Y, Ding SY, Lamed R, Bayer EA. Microbial enzyme systems for biomass conversion: Emerging paradigms. Biofuels. 2010;1:323–41. doi: 10.4155/bfs.09.25. - DOI
-
- De Souza RW. Microbial degradation of lignocellulosic biomass. In: Chandel A, Da Silva, S. Sustainable degradation of lignocellulosic biomass - techniques, applications and commercialization. Brazil: InTech; 2013. doi: 10.5772/54325.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

