Pathogenic cellular role of the p.L104P human cationic trypsinogen variant in chronic pancreatitis
- PMID: 26822915
- PMCID: PMC4824176
- DOI: 10.1152/ajpgi.00444.2015
Pathogenic cellular role of the p.L104P human cationic trypsinogen variant in chronic pancreatitis
Abstract
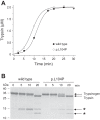
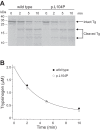
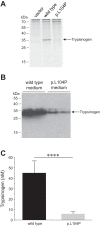
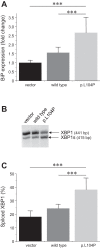
Mutations in the PRSS1 gene encoding human cationic trypsinogen are associated with hereditary and sporadic chronic pancreatitis. High-penetrance PRSS1 mutations found in hereditary pancreatitis alter activation and/or degradation of cationic trypsinogen, thereby promoting intrapancreatic trypsinogen activation. In contrast, a number of rare PRSS1 variants identified in subjects with sporadic chronic pancreatitis cause misfolding and endoplasmic reticulum (ER) stress. Mutation p.L104P is unique among natural PRSS1 variants, since it affects the substrate binding site of trypsin. The aim of the present study was to establish the clinical significance of variant p.L104P through functional analysis. We found that p.L104P trypsin exhibited decreased activity on peptide and protein substrates; however, autoactivation was slightly accelerated. Remarkably, binding of the physiological trypsin inhibitor serine protease inhibitor Kazal type 1 (SPINK1) was decreased by 70-fold. In the presence of the trypsinogen-degrading enzyme chymotrypsin C, mutant p.L104P autoactivated to higher trypsin levels than wild-type trypsinogen. This apparent resistance to degradation was due to slower cleavage at Arg(122) rather than Leu(81) Finally, secretion of mutant p.L104P from transfected cells was markedly reduced due to intracellular retention and aggregation with concomitant elevation of ER stress markers. We conclude that PRSS1 variant p.L104P exhibits a variety of phenotypic changes that can increase risk for chronic pancreatitis. Mutation-induced misfolding and associated ER stress are the dominant effects that support a direct pathogenic role in chronic pancreatitis.
Keywords: autoactivation; chronic pancreatitis; endoplasmic reticulum stress; hereditary pancreatitis; intracellular aggregation; misfolding; trypsinogen activation.
Copyright © 2016 the American Physiological Society.
Figures






Similar articles
-
Functional effects of 13 rare PRSS1 variants presumed to cause chronic pancreatitis.Gut. 2014 Feb;63(2):337-43. doi: 10.1136/gutjnl-2012-304331. Epub 2013 Mar 1. Gut. 2014. PMID: 23455445 Free PMC article.
-
Human cationic trypsinogen (PRSS1) variants and chronic pancreatitis.Am J Physiol Gastrointest Liver Physiol. 2014 Mar;306(6):G466-73. doi: 10.1152/ajpgi.00419.2013. Epub 2014 Jan 23. Am J Physiol Gastrointest Liver Physiol. 2014. PMID: 24458023 Free PMC article. Review.
-
Mesotrypsin Signature Mutation in a Chymotrypsin C (CTRC) Variant Associated with Chronic Pancreatitis.J Biol Chem. 2015 Jul 10;290(28):17282-92. doi: 10.1074/jbc.M114.618439. Epub 2015 May 26. J Biol Chem. 2015. PMID: 26013824 Free PMC article.
-
Misfolding PRSS1 variant p.Ala61Val in a case of suspected intrauterine pancreatitis.Pancreatology. 2025 Feb;25(1):70-81. doi: 10.1016/j.pan.2024.12.013. Epub 2024 Dec 24. Pancreatology. 2025. PMID: 39734120
-
Hereditary chronic pancreatitis.Orphanet J Rare Dis. 2007 Jan 4;2:1. doi: 10.1186/1750-1172-2-1. Orphanet J Rare Dis. 2007. PMID: 17204147 Free PMC article. Review.
Cited by
-
Genetic Risk in Chronic Pancreatitis: The Trypsin-Dependent Pathway.Dig Dis Sci. 2017 Jul;62(7):1692-1701. doi: 10.1007/s10620-017-4601-3. Epub 2017 May 23. Dig Dis Sci. 2017. PMID: 28536777 Free PMC article. Review.
-
ATF6 regulates the development of chronic pancreatitis by inducing p53-mediated apoptosis.Cell Death Dis. 2019 Sep 10;10(9):662. doi: 10.1038/s41419-019-1919-0. Cell Death Dis. 2019. PMID: 31506423 Free PMC article.
-
New insights into the pathways initiating and driving pancreatitis.Curr Opin Gastroenterol. 2016 Sep;32(5):429-435. doi: 10.1097/MOG.0000000000000301. Curr Opin Gastroenterol. 2016. PMID: 27428704 Free PMC article.
-
The 12-Year Experience of the Hungarian Pancreatic Study Group.J Clin Med. 2025 Feb 18;14(4):1362. doi: 10.3390/jcm14041362. J Clin Med. 2025. PMID: 40004893 Free PMC article. Review.
-
Misfolding cationic trypsinogen variant p.L104P causes hereditary pancreatitis.Gut. 2017 Sep;66(9):1727-1728. doi: 10.1136/gutjnl-2016-313451. Epub 2016 Dec 23. Gut. 2017. PMID: 28011893 Free PMC article. No abstract available.
References
-
- Behar DM, Basel-Vanagaite L, Glaser F, Kaplan M, Tzur S, Magal N, Eidlitz-Markus T, Haimi-Cohen Y, Sarig G, Bormans C, Shohat M, Zeharia A. Identification of a novel mutation in the PNLIP gene in two brothers with congenital pancreatic lipase deficiency. J Lipid Res 55: 307–312, 2014. - PMC - PubMed
-
- Bianchini EP, Louvain VB, Marque PE, Juliano MA, Juliano L, Le Bonniec BF. Mapping of the catalytic groove preferences of factor Xa reveals an inadequate selectivity for its macromolecule substrates. J Biol Chem 277: 20527–20534, 2002. - PubMed
-
- Boulling A, Keiles S, Masson E, Chen JM, Férec C. Functional analysis of eight missense mutations in the SPINK1 gene. Pancreas 41: 329–330, 2012. - PubMed
-
- Boulling A, Le Maréchal C, Trouvé P, Raguénès O, Chen JM, Férec C. Functional analysis of pancreatitis-associated missense mutations in the pancreatic secretory trypsin inhibitor (SPINK1) gene. Eur J Hum Genet 15: 936–942, 2007. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

