The evaluation and application of multilocus variable number tandem repeat analysis (MLVA) for the molecular epidemiological study of Salmonella enterica subsp. enterica serovar Enteritidis infection
- PMID: 26823185
- PMCID: PMC4731957
- DOI: 10.1186/s12941-016-0119-3
The evaluation and application of multilocus variable number tandem repeat analysis (MLVA) for the molecular epidemiological study of Salmonella enterica subsp. enterica serovar Enteritidis infection
Abstract
Background: Salmonella enterica subsp. enterica serovar Enteritidis (S. Enteritidis) is one of the most prevalent Salmonella serotypes that cause gastroenteritis worldwide and the most prevalent serotype causing Salmonella infections in China. A rapid molecular typing method with high throughput and good epidemiological discrimination is urgently needed for detecting the outbreaks and finding the source for effective control of S. Enteritidis infections.
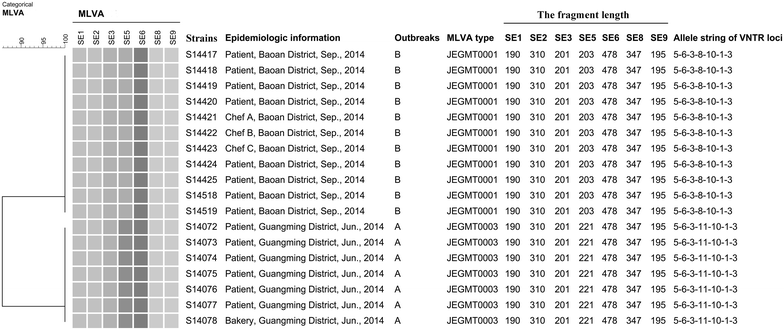
Methods: In this study, 194 strains which included 47 from six outbreaks that were well-characterized epidemiologically were analyzed with pulse field gel electrophoresis (PFGE) and multilocus variable number tandem repeat analysis (MLVA). Seven VNTR loci published by the US Center for Disease Control and Prevention (CDC) were used to evaluate and develop MLVA scheme for S. Enteritidis molecular subtyping by comparing with PFGE, and then MLVA was applied to the suspected outbreaks detection. All S. Enteritidis isolates were analyzed with MLVA to establish a MLVA database in Shenzhen, Guangdong province, China to facilitate the detection of S. Enteritidis infection clusters.
Results: There were 33 MLVA types and 29 PFGE patterns among 147 sporadic isolates. These two measures had Simpson indices of 0.7701 and 0.8043, respectively, which did not differ significantly. Epidemiological concordance was evaluated by typing 47 isolates from six epidemiologically well-characterized outbreaks and it did not differ for PFGE and MLVA. We applied the well established MLVA method to detect two S. Enteritidis foodborne outbreaks and find their sources successfully in 2014. A MLVA database of 491 S. Enteritidis strains isolated from 2004 to 2014 was established for the surveillance of clusters in the future.
Conclusions: MLVA typing of S. Enteritidis would be an effective tool for early warning and epidemiological surveillance of S. Enteritidis infections.
Figures


References
-
- Hong Y, Liu T, Lee MD, Hofacre CL, Maier M, White DG, Ayers S, Wang L, Berghaus R, Maurer J. A rapid screen of broth enrichments for Salmonella enterica serovars enteritidis, Hadar, Heidelberg, and Typhimurium by Using an allelotyping multiplex PCR that targets O- and H-antigen alleles. J Food Prot. 2009;72(10):2198–2201. - PubMed
-
- Sabat AJ, Budimir A, Nashev D, Sa-Leao R, van Dijl J, Laurent F, Grundmann H, Friedrich AW. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 2013;18(4):20380. - PubMed
-
- Deng X, Shariat N, Driebe EM, Roe CC, Tolar B, Trees E, Keim P, Zhang W, Dudley EG, Fields PI, et al. Comparative analysis of subtyping methods against a whole-genome-sequencing standard for Salmonella enterica serotype Enteritidis. J Clin Microbiol. 2015;53(1):212–218. doi: 10.1128/JCM.02332-14. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

