A Biophysical Model of CRISPR/Cas9 Activity for Rational Design of Genome Editing and Gene Regulation
- PMID: 26824432
- PMCID: PMC4732943
- DOI: 10.1371/journal.pcbi.1004724
A Biophysical Model of CRISPR/Cas9 Activity for Rational Design of Genome Editing and Gene Regulation
Abstract
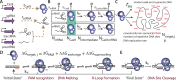
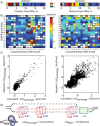
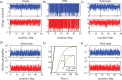
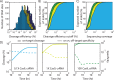
The ability to precisely modify genomes and regulate specific genes will greatly accelerate several medical and engineering applications. The CRISPR/Cas9 (Type II) system binds and cuts DNA using guide RNAs, though the variables that control its on-target and off-target activity remain poorly characterized. Here, we develop and parameterize a system-wide biophysical model of Cas9-based genome editing and gene regulation to predict how changing guide RNA sequences, DNA superhelical densities, Cas9 and crRNA expression levels, organisms and growth conditions, and experimental conditions collectively control the dynamics of dCas9-based binding and Cas9-based cleavage at all DNA sites with both canonical and non-canonical PAMs. We combine statistical thermodynamics and kinetics to model Cas9:crRNA complex formation, diffusion, site selection, reversible R-loop formation, and cleavage, using large amounts of structural, biochemical, expression, and next-generation sequencing data to determine kinetic parameters and develop free energy models. Our results identify DNA supercoiling as a novel mechanism controlling Cas9 binding. Using the model, we predict Cas9 off-target binding frequencies across the lambdaphage and human genomes, and explain why Cas9's off-target activity can be so high. With this improved understanding, we propose several rules for designing experiments for minimizing off-target activity. We also discuss the implications for engineering dCas9-based genetic circuits.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

