Novel Aminoglycoside Resistance Transposons and Transposon-Derived Circular Forms Detected in Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates
- PMID: 26824943
- PMCID: PMC4776018
- DOI: 10.1128/AAC.02143-15
Novel Aminoglycoside Resistance Transposons and Transposon-Derived Circular Forms Detected in Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates
Abstract
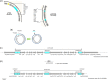
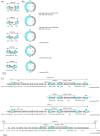
Acinetobacter baumannii has emerged as an important opportunistic pathogen equipped with a growing number of antibiotic resistance genes. Our study investigated the molecular epidemiology and antibiotic resistance features of 28 consecutive carbapenem-resistant clinical isolates of A. baumannii collected throughout Sweden in 2012 and 2013. The isolates mainly belonged to clonal complexes (CCs) with an extensive international distribution, such as CC2 (n = 16) and CC25 (n = 7). Resistance to carbapenems was related to blaOXA-23 (20 isolates), blaOXA-24/40-like (6 isolates), blaOXA-467 (1 isolate), and ISAba1-blaOXA-69 (1 isolate). Ceftazidime resistance was associated with blaPER-7 in the CC25 isolates. Two classical point mutations were responsible for resistance to quinolones in all the isolates. Isolates with high levels of resistance to aminoglycosides carried the 16S rRNA methylase armA gene. The isolates also carried a variety of genes encoding aminoglycoside-modifying enzymes. Several novel structures involved in aminoglycoside resistance were identified, including Tn6279, ΔTn6279, Ab-ST3-aadB, and different assemblies of Tn6020 and TnaphA6. Importantly, a number of circular forms related to the IS26 or ISAba125 composite transposons were detected. The frequent occurrence of these circular forms in the populations of several isolates indicates a potential role of these circular forms in the dissemination of antibiotic resistance genes.
Copyright © 2016 Karah et al.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

