Translational regulation of the mRNA encoding the ubiquitin peptidase USP1 involved in the DNA damage response as a determinant of Cisplatin resistance
- PMID: 26825230
- PMCID: PMC4825832
- DOI: 10.1080/15384101.2015.1120918
Translational regulation of the mRNA encoding the ubiquitin peptidase USP1 involved in the DNA damage response as a determinant of Cisplatin resistance
Erratum in
-
Erratum.Cell Cycle. 2016;15(6):868. doi: 10.1080/23328940.2016.1158033. Cell Cycle. 2016. PMID: 27029531 Free PMC article. No abstract available.
Abstract
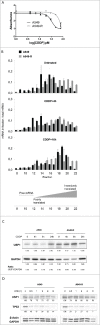
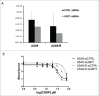
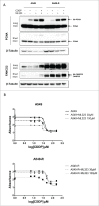
Cisplatin (cis-diaminedichloroplatin (II), CDDP) is part of the standard therapy for a number of solid tumors including Non-Small-Cell Lung Cancer (NSCLC). The initial response observed is in most cases only transient and tumors quickly become refractory to the drug. Tumor cell resistance to CDDP relies on multiple mechanisms, some of which still remain unknown. In search for such mechanisms, we examined the impact of CDDP on mRNA translation in a sensitive and in a matched resistant NSCLC cell line. We identified a set of genes whose mRNAs are differentially translated in CDDP resistant vs. sensitive cells. The translation of the mRNA encoding the Ubiquitin-Specific Peptidase 1 (USP1), a Ubiquitin peptidase with important function in multiple DNA repair pathways, is inhibited by CDDP exposure in the sensitive cells, but not in the resistant cells. This lack of down-regulation of USP1 expression at the translational level plays a primary role in CDDP resistance since inhibition of USP1 expression or activity by siRNA or the small molecule inhibitor ML323, respectively is sufficient to re-sensitize resistant cells to CDDP. We involved the USP1 mRNA translation as a major mechanism of CDDP resistance in NSCLC cells and suggest that USP1 could be evaluated as a candidate predictive marker and as a therapeutic target to overcome CDDP resistance. More generally, our results indicate that analysis of gene expression at the level of mRNA translation is a useful approach to identify new determinants of CDDP resistance.
Keywords: 5′ untranslated region; cisplatin resistance; deubiquitination; lung cancer; monoubiquitination; translation.
Figures

Similar articles
-
A novel mechanism for acquired cisplatin-resistance: suppressed translation of death-associated protein kinase mRNA is insensitive to 5-aza-2'-deoxycitidine and trichostatin in cisplatin-resistant cervical squamous cancer cells.Int J Oncol. 2006 Feb;28(2):497-508. Int J Oncol. 2006. PMID: 16391806
-
Forkhead Box Protein C2 Promotes Epithelial-Mesenchymal Transition, Migration and Invasion in Cisplatin-Resistant Human Ovarian Cancer Cell Line (SKOV3/CDDP).Cell Physiol Biochem. 2016;39(3):1098-110. doi: 10.1159/000447818. Epub 2016 Aug 26. Cell Physiol Biochem. 2016. PMID: 27562816
-
A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses.Nat Chem Biol. 2014 Apr;10(4):298-304. doi: 10.1038/nchembio.1455. Epub 2014 Feb 16. Nat Chem Biol. 2014. PMID: 24531842 Free PMC article.
-
The Role of the Complex USP1/WDR48 in Differentiation and Proliferation Processes in Cancer Stem Cells.Curr Stem Cell Res Ther. 2017;12(5):416-422. doi: 10.2174/1574888X12666170315104013. Curr Stem Cell Res Ther. 2017. PMID: 28302046 Review.
-
USP1 inhibition: A journey from target discovery to clinical translation.Pharmacol Ther. 2025 Jul;271:108865. doi: 10.1016/j.pharmthera.2025.108865. Epub 2025 Apr 22. Pharmacol Ther. 2025. PMID: 40274197 Review.
Cited by
-
Inhibition of Ubiquitin Specific Protease 1 Sensitizes Colorectal Cancer Cells to DNA-Damaging Chemotherapeutics.Front Oncol. 2019 Dec 18;9:1406. doi: 10.3389/fonc.2019.01406. eCollection 2019. Front Oncol. 2019. PMID: 31921663 Free PMC article.
-
Ubiquitin-specific peptidase 1: assessing its role in cancer therapy.Clin Exp Med. 2023 Nov;23(7):2953-2966. doi: 10.1007/s10238-023-01075-4. Epub 2023 Apr 24. Clin Exp Med. 2023. PMID: 37093451 Review.
-
Comprehensive analysis of ubiquitin-specific protease 1 reveals its importance in hepatocellular carcinoma.Cell Prolif. 2020 Oct;53(10):e12908. doi: 10.1111/cpr.12908. Epub 2020 Sep 19. Cell Prolif. 2020. PMID: 32951278 Free PMC article.
-
The USP21/YY1/SNHG16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer.Exp Mol Med. 2020 Jan;52(1):41-55. doi: 10.1038/s12276-019-0356-6. Epub 2020 Jan 20. Exp Mol Med. 2020. PMID: 31956270 Free PMC article.
-
Molecular mechanism of the TP53-MDM2-AR-AKT signalling network regulation by USP12.Oncogene. 2018 Aug;37(34):4679-4691. doi: 10.1038/s41388-018-0283-3. Epub 2018 May 14. Oncogene. 2018. PMID: 29755129
References
-
- Hu L, Wu C, Zhao X, Heist R, Su L, Zhao Y, Han B, Cao S, Chu M, Dai J, et al. . Genome-wide association study of prognosis in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res 2012; 18:5507-14; PMID:22872573; http://dx.doi.org/10.1158/1078-0432.CCR-12-1202 - DOI - PMC - PubMed
-
- Lin M, Stewart DJ, Spitz MR, Hildebrandt MA, Lu C, Lin J, Gu J, Huang M, Lippman SM, Wu X. Genetic variations in the transforming growth factor-β pathway as predictors of survival in advanced non-small cell lung cancer. Carcinogenesis 2011; 32:1050-6; PMID:21515830; http://dx.doi.org/10.1093/carcin/bgr067 - DOI - PMC - PubMed
-
- Li Y, Sun Z, Cunningham JM, Aubry MC, Wampfler JA, Croghan GA, Johnson C, Wu D, Aakre JA, Molina J, et al. . Genetic variations in multiple drug action pathways and survival in advanced stage non-small cell lung cancer treated with chemotherapy. Clin Cancer Res 2011; 17:3830-40; PMID:21636554; http://dx.doi.org/10.1158/1078-0432.CCR-10-2877 - DOI - PMC - PubMed
-
- Wagner KW, Ye Y, Lin J, Vaporciyan AA, Roth JA, Wu X. Genetic variations in epigenetic genes are predictors of recurrence in stage I or II non-small cell lung cancer patients. Clin Cancer Res 2012; 18:585-92; PMID:22252258; http://dx.doi.org/10.1158/1078-0432.CCR-11-2087 - DOI - PMC - PubMed
-
- Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, et al. . Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497:108-12; PMID:23563269; http://dx.doi.org/10.1038/nature12065 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
