Genome Content and Phylogenomics Reveal both Ancestral and Lateral Evolutionary Pathways in Plant-Pathogenic Streptomyces Species
- PMID: 26826232
- PMCID: PMC4807529
- DOI: 10.1128/AEM.03504-15
Genome Content and Phylogenomics Reveal both Ancestral and Lateral Evolutionary Pathways in Plant-Pathogenic Streptomyces Species
Abstract
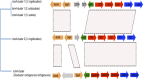
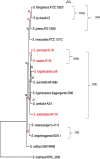
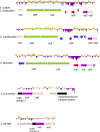
Streptomyces spp. are highly differentiated actinomycetes with large, linear chromosomes that encode an arsenal of biologically active molecules and catabolic enzymes. Members of this genus are well equipped for life in nutrient-limited environments and are common soil saprophytes. Out of the hundreds of species in the genus Streptomyces, a small group has evolved the ability to infect plants. The recent availability of Streptomyces genome sequences, including four genomes of pathogenic species, provided an opportunity to characterize the gene content specific to these pathogens and to study phylogenetic relationships among them. Genome sequencing, comparative genomics, and phylogenetic analysis enabled us to discriminate pathogenic from saprophytic Streptomyces strains; moreover, we calculated that the pathogen-specific genome contains 4,662 orthologs. Phylogenetic reconstruction suggested that Streptomyces scabies and S. ipomoeae share an ancestor but that their biosynthetic clusters encoding the required virulence factor thaxtomin have diverged. In contrast, S. turgidiscabies and S. acidiscabies, two relatively unrelated pathogens, possess highly similar thaxtomin biosynthesis clusters, which suggests that the acquisition of these genes was through lateral gene transfer.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang C-H, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream M-A, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi: 10.1038/417141a. - DOI - PubMed
-
- Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U S A 98:12215–12220. doi: 10.1073/pnas.211433198. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

