The epigenetic modifier DNMT3A is necessary for proper otic placode formation
- PMID: 26826496
- PMCID: PMC4783191
- DOI: 10.1016/j.ydbio.2016.01.034
The epigenetic modifier DNMT3A is necessary for proper otic placode formation
Abstract
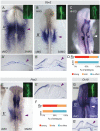
Cranial placodes are thickenings in the ectoderm that give rise to sensory organs and peripheral ganglia of the vertebrate head. At gastrula and neurula stages, placodal precursors are intermingled in the neural plate border with future neural and neural crest cells. Here, we show that the epigenetic modifier, DNA methyl transferase (DNMT) 3A, expressed in the neural plate border region, influences development of the otic placode which will contribute to the ear. DNMT3A is expressed in the presumptive otic region at gastrula through neurula stages and later in the otic placode itself. Whereas neural plate border and non-neural ectoderm markers Erni, Dlx5, Msx1 and Six1 are unaltered, DNMT3A loss of function leads to early reduction in the expression of the key otic placode specifier genes Pax2 and Gbx2 and later otic markers Sox10 and Soho1. Reduction of Gbx2 was first observed at HH7, well before loss of other otic markers. Later, this translates to significant reduction in the size of the otic vesicle. Based on these results, we propose that DNMT3A is important for enabling the activation of Gbx2 expression, necessary for normal development of the inner ear.
Keywords: DNMT3A; Gbx2; Otic; Pax2; Placode.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Foxi3 is necessary for the induction of the chick otic placode in response to FGF signaling.Dev Biol. 2014 Jul 15;391(2):158-69. doi: 10.1016/j.ydbio.2014.04.014. Epub 2014 Apr 26. Dev Biol. 2014. PMID: 24780628 Free PMC article.
-
Six1 and Irx1 have reciprocal interactions during cranial placode and otic vesicle formation.Dev Biol. 2019 Feb 1;446(1):68-79. doi: 10.1016/j.ydbio.2018.12.003. Epub 2018 Dec 6. Dev Biol. 2019. PMID: 30529252 Free PMC article.
-
Extensive cell movements accompany formation of the otic placode.Dev Biol. 2002 Sep 15;249(2):237-54. doi: 10.1006/dbio.2002.0739. Dev Biol. 2002. PMID: 12221004
-
Early embryonic specification of vertebrate cranial placodes.Wiley Interdiscip Rev Dev Biol. 2014 Sep-Oct;3(5):349-63. doi: 10.1002/wdev.142. Epub 2014 Jul 2. Wiley Interdiscip Rev Dev Biol. 2014. PMID: 25124756 Review.
-
Changing shape and shaping change: Inducing the inner ear.Semin Cell Dev Biol. 2017 May;65:39-46. doi: 10.1016/j.semcdb.2016.10.006. Epub 2016 Oct 29. Semin Cell Dev Biol. 2017. PMID: 27989562 Review.
Cited by
-
DNA Methyltransferases: From Evolution to Clinical Applications.Int J Mol Sci. 2022 Aug 12;23(16):8994. doi: 10.3390/ijms23168994. Int J Mol Sci. 2022. PMID: 36012258 Free PMC article. Review.
-
Dynamic transcriptional signature and cell fate analysis reveals plasticity of individual neural plate border cells.Elife. 2017 Mar 29;6:e21620. doi: 10.7554/eLife.21620. Elife. 2017. PMID: 28355135 Free PMC article.
-
Lack of Methylation Changes in GJB2 and RB1 Non-coding Regions of Cochlear Implant Patients with Sensorineural Hearing Loss.Acta Med Philipp. 2023 Sep 28;57(9):116-120. doi: 10.47895/amp.v57i9.5200. Acta Med Philipp. 2023. PMID: 37990697 Free PMC article.
-
Role of microRNAs in inner ear development and hearing loss.Gene. 2019 Feb 20;686:49-55. doi: 10.1016/j.gene.2018.10.075. Epub 2018 Oct 31. Gene. 2019. PMID: 30389561 Free PMC article. Review.
-
Sensing External and Self-Motion with Hair Cells: A Comparison of the Lateral Line and Vestibular Systems from a Developmental and Evolutionary Perspective.Brain Behav Evol. 2017;90(2):98-116. doi: 10.1159/000456646. Epub 2017 Oct 9. Brain Behav Evol. 2017. PMID: 28988233 Free PMC article. Review.
References
-
- Acloque H, Wilkinson DG, Nieto MA. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods in cell biology. 2008;87:169–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

