Structural characterisation of human galectin-4 N-terminal carbohydrate recognition domain in complex with glycerol, lactose, 3'-sulfo-lactose, and 2'-fucosyllactose
- PMID: 26828567
- PMCID: PMC4734333
- DOI: 10.1038/srep20289
Structural characterisation of human galectin-4 N-terminal carbohydrate recognition domain in complex with glycerol, lactose, 3'-sulfo-lactose, and 2'-fucosyllactose
Abstract
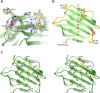
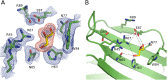
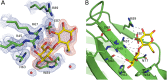
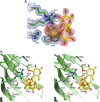
Galectin-4 is a tandem-repeat galectin with two distinct carbohydrate recognition domains (CRD). Galectin-4 is expressed mainly in the alimentary tract and is proposed to function as a lipid raft and adherens junction stabilizer by its glycan cross-linking capacity. Galectin-4 plays divergent roles in cancer and inflammatory conditions, either promoting or inhibiting each disease progression, depending on the specific pathological condition. The study of galectin-4's ligand-binding profile may help decipher its roles under specific conditions. Here we present the X-ray structures of human galectin-4 N-terminal CRD (galectin-4N) bound to different saccharide ligands. Galectin-4's overall fold and its core interactions to lactose are similar to other galectin CRDs. Galectin-4N recognises the sulfate cap of 3'-sulfated glycans by a weak interaction through Arg45 and two water-mediated hydrogen bonds via Trp84 and Asn49. When galectin-4N interacts with the H-antigen mimic, 2'-fucosyllactose, an interaction is formed between the ring oxygen of fucose and Arg45. The extended binding site of galectin-4N may not be well suited to the A/B-antigen determinants, α-GalNAc/α-Gal, specifically due to clashes with residue Phe47. Overall, galectin-4N favours sulfated glycans whilst galectin-4C prefers blood group determinants. However, the two CRDs of galectin-4 can, to a less extent, recognise each other's ligands.
Figures

Similar articles
-
Structural characterization of human galectin-4 C-terminal domain: elucidating the molecular basis for recognition of glycosphingolipids, sulfated saccharides and blood group antigens.FEBS J. 2015 Sep;282(17):3348-67. doi: 10.1111/febs.13348. Epub 2015 Jul 14. FEBS J. 2015. PMID: 26077389
-
Structure of the mouse galectin-4 N-terminal carbohydrate-recognition domain reveals the mechanism of oligosaccharide recognition.Acta Crystallogr D Biol Crystallogr. 2011 Mar;67(Pt 3):204-11. doi: 10.1107/S0907444911004082. Epub 2011 Feb 15. Acta Crystallogr D Biol Crystallogr. 2011. PMID: 21358051
-
Crystallization and preliminary X-ray diffraction analysis of mouse galectin-4 N-terminal carbohydrate recognition domain in complex with lactose.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 Jul 1;64(Pt 7):665-7. doi: 10.1107/S1744309108017405. Epub 2008 Jun 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18607104 Free PMC article.
-
Galectin-4 in normal tissues and cancer.Glycoconj J. 2004;20(4):247-55. doi: 10.1023/B:GLYC.0000025819.54723.a0. Glycoconj J. 2004. PMID: 15115909 Review.
-
The role of galectin-4 in physiology and diseases.Protein Cell. 2016 May;7(5):314-24. doi: 10.1007/s13238-016-0262-9. Epub 2016 Mar 26. Protein Cell. 2016. PMID: 27017379 Free PMC article. Review.
Cited by
-
A brief history of galectin evolution.Front Immunol. 2023 Jun 29;14:1147356. doi: 10.3389/fimmu.2023.1147356. eCollection 2023. Front Immunol. 2023. PMID: 37457740 Free PMC article.
-
Binding of Glycerol to Human Galectin-7 Expands Stability and Modulates Its Functions.Int J Mol Sci. 2022 Oct 14;23(20):12318. doi: 10.3390/ijms232012318. Int J Mol Sci. 2022. PMID: 36293173 Free PMC article.
-
Galectin inhibitors and nanoparticles as a novel therapeutic strategy for glioblastoma multiforme.Am J Cancer Res. 2024 Feb 15;14(2):774-795. doi: 10.62347/MKIV1986. eCollection 2024. Am J Cancer Res. 2024. PMID: 38455415 Free PMC article. Review.
-
Chicken GRIFIN: Structural characterization in crystals and in solution.Biochimie. 2018 Mar;146:127-138. doi: 10.1016/j.biochi.2017.12.003. Epub 2017 Dec 15. Biochimie. 2018. PMID: 29248541 Free PMC article.
-
Modulation of the Gal-9/TIM-3 Immune Checkpoint with α-Lactose. Does Anomery of Lactose Matter?Cancers (Basel). 2021 Dec 18;13(24):6365. doi: 10.3390/cancers13246365. Cancers (Basel). 2021. PMID: 34944985 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

