PSAMM: A Portable System for the Analysis of Metabolic Models
- PMID: 26828591
- PMCID: PMC4734835
- DOI: 10.1371/journal.pcbi.1004732
PSAMM: A Portable System for the Analysis of Metabolic Models
Abstract
The genome-scale models of metabolic networks have been broadly applied in phenotype prediction, evolutionary reconstruction, community functional analysis, and metabolic engineering. Despite the development of tools that support individual steps along the modeling procedure, it is still difficult to associate mathematical simulation results with the annotation and biological interpretation of metabolic models. In order to solve this problem, here we developed a Portable System for the Analysis of Metabolic Models (PSAMM), a new open-source software package that supports the integration of heterogeneous metadata in model annotations and provides a user-friendly interface for the analysis of metabolic models. PSAMM is independent of paid software environments like MATLAB, and all its dependencies are freely available for academic users. Compared to existing tools, PSAMM significantly reduced the running time of constraint-based analysis and enabled flexible settings of simulation parameters using simple one-line commands. The integration of heterogeneous, model-specific annotation information in PSAMM is achieved with a novel format of YAML-based model representation, which has several advantages, such as providing a modular organization of model components and simulation settings, enabling model version tracking, and permitting the integration of multiple simulation problems. PSAMM also includes a number of quality checking procedures to examine stoichiometric balance and to identify blocked reactions. Applying PSAMM to 57 models collected from current literature, we demonstrated how the software can be used for managing and simulating metabolic models. We identified a number of common inconsistencies in existing models and constructed an updated model repository to document the resolution of these inconsistencies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

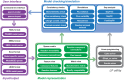

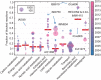

Similar articles
-
Using PSAMM for the Curation and Analysis of Genome-Scale Metabolic Models.Methods Mol Biol. 2018;1716:131-150. doi: 10.1007/978-1-4939-7528-0_6. Methods Mol Biol. 2018. PMID: 29222752
-
ReMatch: a web-based tool to construct, store and share stoichiometric metabolic models with carbon maps for metabolic flux analysis.J Integr Bioinform. 2008 Aug 25;5(2). doi: 10.2390/biecoll-jib-2008-102. J Integr Bioinform. 2008. PMID: 20134058
-
ModelExplorer - software for visual inspection and inconsistency correction of genome-scale metabolic reconstructions.BMC Bioinformatics. 2019 Jan 28;20(1):56. doi: 10.1186/s12859-019-2615-x. BMC Bioinformatics. 2019. PMID: 30691403 Free PMC article.
-
Use of CellNetAnalyzer in biotechnology and metabolic engineering.J Biotechnol. 2017 Nov 10;261:221-228. doi: 10.1016/j.jbiotec.2017.05.001. Epub 2017 May 10. J Biotechnol. 2017. PMID: 28499817 Review.
-
Coupling Fluxes, Enzymes, and Regulation in Genome-Scale Metabolic Models.Methods Mol Biol. 2018;1716:337-351. doi: 10.1007/978-1-4939-7528-0_15. Methods Mol Biol. 2018. PMID: 29222761 Review.
Cited by
-
Metabolic engineering of Caldicellulosiruptor bescii for 2,3-butanediol production from unpretreated lignocellulosic biomass and metabolic strategies for improving yields and titers.Appl Environ Microbiol. 2024 Jan 24;90(1):e0195123. doi: 10.1128/aem.01951-23. Epub 2023 Dec 22. Appl Environ Microbiol. 2024. PMID: 38131671 Free PMC article.
-
Identification of a deep-branching thermophilic clade sheds light on early bacterial evolution.Nat Commun. 2023 Jul 19;14(1):4354. doi: 10.1038/s41467-023-39960-x. Nat Commun. 2023. PMID: 37468486 Free PMC article.
-
The genome sequence of the grape phylloxera provides insights into the evolution, adaptation, and invasion routes of an iconic pest.BMC Biol. 2020 Jul 23;18(1):90. doi: 10.1186/s12915-020-00820-5. BMC Biol. 2020. PMID: 32698880 Free PMC article.
-
Microbiome Analysis Reveals Diversity and Function of Mollicutes Associated with the Eastern Oyster, Crassostrea virginica.mSphere. 2021 May 12;6(3):e00227-21. doi: 10.1128/mSphere.00227-21. mSphere. 2021. PMID: 33980678 Free PMC article.
-
Isolating structural errors in reaction networks in systems biology.Bioinformatics. 2021 Apr 20;37(3):388-395. doi: 10.1093/bioinformatics/btaa720. Bioinformatics. 2021. PMID: 32790862 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

