The Functional Unit of Neisseria meningitidis 3-Deoxy-ᴅ-Arabino-Heptulosonate 7-Phosphate Synthase Is Dimeric
- PMID: 26828675
- PMCID: PMC4735112
- DOI: 10.1371/journal.pone.0145187
The Functional Unit of Neisseria meningitidis 3-Deoxy-ᴅ-Arabino-Heptulosonate 7-Phosphate Synthase Is Dimeric
Abstract
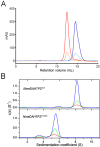
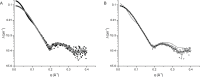
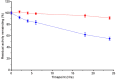
Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (NmeDAH7PS) adopts a homotetrameric structure consisting of an extensive and a less extensive interface. Perturbation of the less extensive interface through a single mutation of a salt bridge (Arg126-Glu27) formed at the tetramer interface of all chains resulted in a dimeric DAH7PS in solution, as determined by small angle X-ray scattering, analytical ultracentrifugation and analytical size-exclusion chromatography. The dimeric NmeDAH7PSR126S variant was shown to be catalytically active in the aldol-like condensation reaction between D-erythrose 4-phosphate and phosphoenolpyruvate, and allosterically inhibited by L-phenylalanine to the same extent as the wild-type enzyme. The dimeric NmeDAH7PSR126S variant exhibited a slight reduction in thermal stability by differential scanning calorimetry experiments and a slow loss of activity over time compared to the wild-type enzyme. Although NmeDAH7PSR126S crystallised as a tetramer, like the wild-type enzyme, structural asymmetry at the less extensive interface was observed consistent with its destabilisation. The tetrameric association enabled by this Arg126-Glu27 salt-bridge appears to contribute solely to the stability of the protein, ultimately revealing that the functional unit of NmeDAH7PS is dimeric.
Conflict of interest statement
Figures



Similar articles
-
Destabilization of the homotetrameric assembly of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from the hyperthermophile Pyrococcus furiosus enhances enzymatic activity.J Mol Biol. 2014 Feb 6;426(3):656-73. doi: 10.1016/j.jmb.2013.11.008. Epub 2013 Nov 14. J Mol Biol. 2014. PMID: 24239948
-
Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.Protein Sci. 2013 Aug;22(8):1087-99. doi: 10.1002/pro.2293. Epub 2013 Jun 27. Protein Sci. 2013. PMID: 23754471 Free PMC article.
-
Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase - The importance of accommodating the active site water.Bioorg Chem. 2014 Dec;57:242-250. doi: 10.1016/j.bioorg.2014.08.003. Epub 2014 Aug 27. Bioorg Chem. 2014. PMID: 25245459
-
Substrate ambiguity and crystal structure of Pyrococcus furiosus 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase: an ancestral 3-deoxyald-2-ulosonate-phosphate synthase?Biochemistry. 2005 Sep 13;44(36):11950-62. doi: 10.1021/bi050577z. Biochemistry. 2005. PMID: 16142893
-
Diverse allosteric componentry and mechanisms control entry into aromatic metabolite biosynthesis.Curr Opin Struct Biol. 2020 Dec;65:159-167. doi: 10.1016/j.sbi.2020.06.015. Epub 2020 Jul 30. Curr Opin Struct Biol. 2020. PMID: 32739636 Review.
Cited by
-
Domain cross-talk within a bifunctional enzyme provides catalytic and allosteric functionality in the biosynthesis of aromatic amino acids.J Biol Chem. 2019 Mar 29;294(13):4828-4842. doi: 10.1074/jbc.RA118.005220. Epub 2019 Jan 22. J Biol Chem. 2019. PMID: 30670586 Free PMC article.
References
-
- Bentley R. The Shikimate Pathway—A Metabolic Tree with Many Branches. Crit Rev Biochem Mol Biol. 1990;25: 307–384. - PubMed
-
- Webby C, Baker H, Lott S, Baker E, Parker E. The Structure of 3-Deoxy-D-arabino-heptulosonate 7-phosphate Synthase from Mycobacterium tuberculosis Reveals a Common Catalytic Scaffold and Ancestry for Type I and Type II Enzymes. J Mol Biol. 2005;354: 927–939. - PubMed
-
- Wu J, Woodard RW. New insights into the evolutionary links relating to the 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase subfamilies. J Biol Chem. 2006;281: 4042–4048. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

