Arabidopsis WRKY6 Transcription Factor Acts as a Positive Regulator of Abscisic Acid Signaling during Seed Germination and Early Seedling Development
- PMID: 26829043
- PMCID: PMC4734665
- DOI: 10.1371/journal.pgen.1005833
Arabidopsis WRKY6 Transcription Factor Acts as a Positive Regulator of Abscisic Acid Signaling during Seed Germination and Early Seedling Development
Erratum in
-
Correction: Arabidopsis WRKY6 Transcription Factor Acts as a Positive Regulator of Abscisic Acid Signaling during Seed Germination and Early Seedling Development.PLoS Genet. 2019 Mar 6;15(3):e1008032. doi: 10.1371/journal.pgen.1008032. eCollection 2019 Mar. PLoS Genet. 2019. PMID: 30840622 Free PMC article.
Abstract
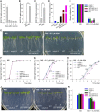
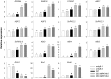
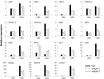
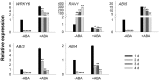
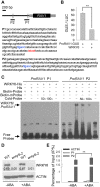
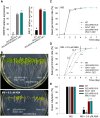
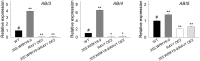
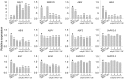
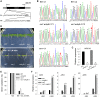
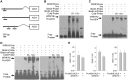
The phytohormone abscisic acid (ABA) plays important roles during seed germination and early seedling development. Here, we characterized the function of the Arabidopsis WRKY6 transcription factor in ABA signaling. The transcript of WRKY6 was repressed during seed germination and early seedling development, and induced by exogenous ABA. The wrky6-1 and wrky6-2 mutants were ABA insensitive, whereas WRKY6-overexpressing lines showed ABA-hypersensitive phenotypes during seed germination and early seedling development. The expression of RAV1 was suppressed in the WRKY6-overexpressing lines and elevated in the wrky6 mutants, and the expression of ABI3, ABI4, and ABI5, which was directly down-regulated by RAV1, was enhanced in the WRKY6-overexpressing lines and repressed in the wrky6 mutants. Electrophoretic mobility shift and chromatin immunoprecipitation assays showed that WRKY6 could bind to the RAV1 promoter in vitro and in vivo. Overexpression of RAV1 in WRKY6-overexpressing lines abolished their ABA-hypersensitive phenotypes, and the rav1 wrky6-2 double mutant showed an ABA-hypersensitive phenotype, similar to rav1 mutant. Together, the results demonstrated that the Arabidopsis WRKY6 transcription factor played important roles in ABA signaling by directly down-regulating RAV1 expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

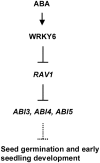
Similar articles
-
Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development.Plant J. 2014 Nov;80(4):654-68. doi: 10.1111/tpj.12670. Plant J. 2014. PMID: 25231920
-
ARABIDOPSIS NITRATE REGULATED 1 acts as a negative modulator of seed germination by activating ABI3 expression.New Phytol. 2020 Jan;225(2):835-847. doi: 10.1111/nph.16172. Epub 2019 Oct 14. New Phytol. 2020. PMID: 31491809
-
Abscisic Acid Modulates Seed Germination via ABA INSENSITIVE5-Mediated PHOSPHATE1.Plant Physiol. 2017 Dec;175(4):1661-1668. doi: 10.1104/pp.17.00164. Epub 2017 Oct 31. Plant Physiol. 2017. PMID: 29089393 Free PMC article.
-
Light and abscisic acid interplay in early seedling development.New Phytol. 2021 Jan;229(2):763-769. doi: 10.1111/nph.16963. Epub 2020 Oct 25. New Phytol. 2021. PMID: 32984965 Review.
-
Multifaceted Signaling Networks Mediated by Abscisic Acid Insensitive 4.Plant Commun. 2020 Mar 7;1(3):100040. doi: 10.1016/j.xplc.2020.100040. eCollection 2020 May 11. Plant Commun. 2020. PMID: 33367237 Free PMC article. Review.
Cited by
-
The Ubiquitin E3 Ligase PRU2 Modulates Phosphate Uptake in Arabidopsis.Int J Mol Sci. 2022 Feb 18;23(4):2273. doi: 10.3390/ijms23042273. Int J Mol Sci. 2022. PMID: 35216388 Free PMC article.
-
Seed Longevity in Legumes: Deeper Insights Into Mechanisms and Molecular Perspectives.Front Plant Sci. 2022 Jul 27;13:918206. doi: 10.3389/fpls.2022.918206. eCollection 2022. Front Plant Sci. 2022. PMID: 35968115 Free PMC article.
-
Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs.BMC Plant Biol. 2021 Mar 16;21(1):137. doi: 10.1186/s12870-021-02907-9. BMC Plant Biol. 2021. PMID: 33726681 Free PMC article.
-
Genomic identification and expression profiling of WRKY genes in alfalfa (Medicago sativa) elucidate their responsiveness to seed vigor.BMC Plant Biol. 2023 Nov 16;23(1):568. doi: 10.1186/s12870-023-04597-x. BMC Plant Biol. 2023. PMID: 37968658 Free PMC article.
-
Genome-Wide Identification and Characterization of CDPK Family Reveal Their Involvements in Growth and Development and Abiotic Stress in Sweet Potato and Its Two Diploid Relatives.Int J Mol Sci. 2022 Mar 13;23(6):3088. doi: 10.3390/ijms23063088. Int J Mol Sci. 2022. PMID: 35328509 Free PMC article.
References
-
- Verslues PE, Zhu JK. New developments in abscisic acid perception and metabolism. Curr Opin Plant Biol. 2007;27: 7781–7790. - PubMed
-
- Ali-Rachedi S, Bouinot D, Wagner MH, Bonnet M, Sotta B, Grappin P, et al. Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta. 2004;219: 479–488. - PubMed
-
- Gubler F, Millar AA, Jacobsen JV. Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol. 2005;8: 183–187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

