Methylation Markers for the Identification of Body Fluids and Tissues from Forensic Trace Evidence
- PMID: 26829227
- PMCID: PMC4734623
- DOI: 10.1371/journal.pone.0147973
Methylation Markers for the Identification of Body Fluids and Tissues from Forensic Trace Evidence
Erratum in
-
Correction: Methylation Markers for the Identification of Body Fluids and Tissues from Forensic Trace Evidence.PLoS One. 2016 May 24;11(5):e0156472. doi: 10.1371/journal.pone.0156472. eCollection 2016. PLoS One. 2016. PMID: 27219355 Free PMC article.
Abstract
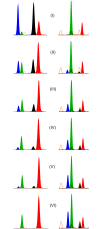
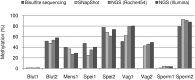
The identification of body fluids is an essential tool for clarifying the course of events at a criminal site. The analytical problem is the fact that the biological material has been very often exposed to detrimental exogenous influences. Thereby, the molecular substrates used for the identification of the traces may become degraded. So far, most protocols utilize cell specific proteins or RNAs. Instead of measuring these more sensitive compounds this paper describes the application of the differential DNA-methylation. As a result of two genome wide screenings with the Illumina HumanMethylation BeadChips 27 and 450k we identified 150 candidate loci revealing differential methylation with regard to the body fluids venous blood, menstrual blood, vaginal fluid, saliva and sperm. Among them we selected 9 loci as the most promising markers. For the final determination of the methylation degree we applied the SNuPE-method. Because the degree of methylation might be modified by various endogenous and exogenous factors, we tested each marker with approximately 100 samples of each target fluid in a validation study. The stability of the detection procedure is proved in various simulated forensic surroundings according to standardized conditions. We studied the potential influence of 12 relatively common tumors on the methylation of the 9 markers. For this purpose the target fluids of 34 patients have been analysed. Only the cervix carcinoma might have an remarkable effect because impairing the signal of both vaginal markers. Using the Illumina MiSeq device we tested the potential influence of cis acting sequence variants on the methylation degree of the 9 markers in the specific body fluid DNA of 50 individuals. For 4 marker loci we observed such an influence either by sole SNPs or haplotypes. The identification of each target fluid is possible in arbitrary mixtures with the remaining four body fluids. The sensitivity of the individual body fluid tests is in the same range as for the forensic STR-analysis. It is the first forensic body fluid protocol which considers the exogenic and endogenic parameters potentially interfering with the true results.
Conflict of interest statement
Figures




Similar articles
-
Identification of body fluid-specific DNA methylation markers for use in forensic science.Forensic Sci Int Genet. 2014 Nov;13:147-53. doi: 10.1016/j.fsigen.2014.07.011. Epub 2014 Jul 27. Forensic Sci Int Genet. 2014. PMID: 25128690
-
Genome-wide methylation profiling and a multiplex construction for the identification of body fluids using epigenetic markers.Forensic Sci Int Genet. 2015 Jul;17:17-24. doi: 10.1016/j.fsigen.2015.03.002. Epub 2015 Mar 12. Forensic Sci Int Genet. 2015. PMID: 25796047
-
DNA methylation profiling for a confirmatory test for blood, saliva, semen, vaginal fluid and menstrual blood.Forensic Sci Int Genet. 2016 Sep;24:75-82. doi: 10.1016/j.fsigen.2016.06.007. Epub 2016 Jun 14. Forensic Sci Int Genet. 2016. PMID: 27344518
-
Molecular approaches for forensic cell type identification: On mRNA, miRNA, DNA methylation and microbial markers.Forensic Sci Int Genet. 2015 Sep;18:21-32. doi: 10.1016/j.fsigen.2014.11.015. Epub 2014 Nov 23. Forensic Sci Int Genet. 2015. PMID: 25488609 Review.
-
Forensic DNA methylation profiling--potential opportunities and challenges.Forensic Sci Int Genet. 2013 Sep;7(5):499-507. doi: 10.1016/j.fsigen.2013.05.004. Epub 2013 Jun 28. Forensic Sci Int Genet. 2013. PMID: 23948320 Review.
Cited by
-
Forensic Epigenetic Analysis: The Path Ahead.Med Princ Pract. 2019;28(4):301-308. doi: 10.1159/000499496. Epub 2019 Mar 12. Med Princ Pract. 2019. PMID: 30893697 Free PMC article. Review.
-
From forensic epigenetics to forensic epigenomics: broadening DNA investigative intelligence.Genome Biol. 2017 Dec 21;18(1):238. doi: 10.1186/s13059-017-1373-1. Genome Biol. 2017. PMID: 29268765 Free PMC article.
-
Forensic age prediction for saliva samples using methylation-sensitive high resolution melting: exploratory application for cigarette butts.Sci Rep. 2017 Sep 5;7(1):10444. doi: 10.1038/s41598-017-10752-w. Sci Rep. 2017. PMID: 28874809 Free PMC article.
-
Characterization of DNA methylation-based markers for human body fluid identification in forensics: a critical review.Int J Legal Med. 2020 Jan;134(1):1-20. doi: 10.1007/s00414-019-02181-3. Epub 2019 Nov 12. Int J Legal Med. 2020. PMID: 31713682 Review.
-
A new approach to epigenome-wide discovery of non-invasive methylation biomarkers for colorectal cancer screening in circulating cell-free DNA using pooled samples.Clin Epigenetics. 2018 Apr 16;10:53. doi: 10.1186/s13148-018-0487-y. eCollection 2018. Clin Epigenetics. 2018. PMID: 29686738 Free PMC article.
References
-
- Santucci KA, Nelson DG, McQuillen KK, Duffy SJ, Linakis JG. Wood's lamp utility in the identification of semen. Pediatrics. 1999. December;104(6):1342–4. - PubMed
-
- James SH, Nordby JJ. Identification of bi-ological fluids and stains; Forensic science: an introduction to scientific and investigative techniques. 2nd ed Boca Raton: CRC Press, USA; 2003.
-
- Allen SM. An enzyme linked immunosorbent assay (ELISA) for detection of seminal fluid using a monoclonal antibody to prostatic acid phosphatase. J Immunoassay. 1995. August;16(3):297–308. - PubMed
-
- Sato I, Barni F, Yoshiike M, Rapone C, Berti A, Nakaki S, et al. Applicability of Nanotrap Sg as a semen detection kit before male-specific DNA profiling in sexual assaults. Int J Legal Med. 2007. July;121(4):315–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

