The Genetic Architecture of Murine Glutathione Transferases
- PMID: 26829228
- PMCID: PMC4734686
- DOI: 10.1371/journal.pone.0148230
The Genetic Architecture of Murine Glutathione Transferases
Abstract
Glutathione S-transferase (GST) genes play a protective role against oxidative stress and may influence disease risk and drug pharmacokinetics. In this study, massive multiscalar trait profiling across a large population of mice derived from a cross between C57BL/6J (B6) and DBA2/J (D2)--the BXD family--was combined with linkage and bioinformatic analyses to characterize mechanisms controlling GST expression and to identify downstream consequences of this variation. Similar to humans, mice show a wide range in expression of GST family members. Variation in the expression of Gsta4, Gstt2, Gstz1, Gsto1, and Mgst3 is modulated by local expression QTLs (eQTLs) in several tissues. Higher expression of Gsto1 in brain and liver of BXD strains is strongly associated (P < 0.01) with inheritance of the B6 parental allele whereas higher expression of Gsta4 and Mgst3 in brain and liver, and Gstt2 and Gstz1 in brain is strongly associated with inheritance of the D2 parental allele. Allele-specific assays confirmed that expression of Gsto1, Gsta4, and Mgst3 are modulated by sequence variants within or near each gene locus. We exploited this endogenous variation to identify coexpression networks and downstream targets in mouse and human. Through a combined systems genetics approach, we provide new insight into the biological role of naturally occurring variants in GST genes.
Conflict of interest statement
Figures
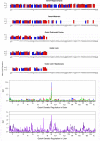
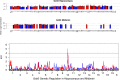
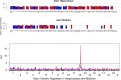
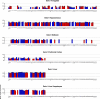
Similar articles
-
Expression quantitative trait loci mapping identifies new genetic models of glutathione S-transferase variation.Drug Metab Dispos. 2009 Jun;37(6):1269-76. doi: 10.1124/dmd.109.026856. Epub 2009 Mar 26. Drug Metab Dispos. 2009. PMID: 19324942
-
Genetic and epigenetic regulation and expression signatures of glutathione S-transferases in developing mouse liver.Toxicol Sci. 2010 Jul;116(1):32-43. doi: 10.1093/toxsci/kfq115. Epub 2010 Apr 15. Toxicol Sci. 2010. PMID: 20395309 Free PMC article.
-
Using gene expression databases for classical trait QTL candidate gene discovery in the BXD recombinant inbred genetic reference population: mouse forebrain weight.BMC Genomics. 2008 Sep 25;9:444. doi: 10.1186/1471-2164-9-444. BMC Genomics. 2008. PMID: 18817551 Free PMC article.
-
Pi-class glutathione S-transferase: regulation and function.Chem Biol Interact. 1998 Apr 24;111-112:69-82. doi: 10.1016/s0009-2797(97)00176-2. Chem Biol Interact. 1998. PMID: 9679544 Review.
-
Characterization of the omega class of glutathione transferases.Methods Enzymol. 2005;401:78-99. doi: 10.1016/S0076-6879(05)01005-0. Methods Enzymol. 2005. PMID: 16399380 Review.
Cited by
-
Lipid droplet-associated kinase STK25 regulates peroxisomal activity and metabolic stress response in steatotic liver.J Lipid Res. 2020 Feb;61(2):178-191. doi: 10.1194/jlr.RA119000316. Epub 2019 Dec 19. J Lipid Res. 2020. PMID: 31857389 Free PMC article.
-
Single-cell allele-specific expression analysis reveals dynamic and cell-type-specific regulatory effects.Nat Commun. 2023 Oct 9;14(1):6317. doi: 10.1038/s41467-023-42016-9. Nat Commun. 2023. PMID: 37813843 Free PMC article.
-
Integrated metabolomics and transcriptomics reveal the anti-aging effect of melanin from Sepiella maindroni ink (MSMI) on D-galactose-induced aging mice.Aging (Albany NY). 2021 Apr 21;13(8):11889-11906. doi: 10.18632/aging.202890. Epub 2021 Apr 21. Aging (Albany NY). 2021. PMID: 33952720 Free PMC article.
-
UBL3 Interaction with α-Synuclein Is Downregulated by Silencing MGST3.Biomedicines. 2023 Sep 8;11(9):2491. doi: 10.3390/biomedicines11092491. Biomedicines. 2023. PMID: 37760932 Free PMC article.
-
The Notch Ligand Jagged1 Is Required for the Formation, Maintenance, and Survival of Hensen's Cells in the Mouse Cochlea.J Neurosci. 2020 Dec 2;40(49):9401-9413. doi: 10.1523/JNEUROSCI.1192-20.2020. Epub 2020 Oct 30. J Neurosci. 2020. PMID: 33127852 Free PMC article.
References
-
- Hayes JD, Flanagan JU, Jowsey IR (2005) Glutathione transferases. Annu Rev Pharmacol Toxicol 45: 51–88. - PubMed
-
- Mannervik B, Danielson UH (1988) Glutathione transferases—structure and catalytic activity. CRC Crit Rev Biochem 23: 283–337. - PubMed
-
- Board PG (1981) Transport of glutathione S-conjugate from human erythrocytes. FEBS Lett 124: 163–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

