Haplotyping germline and cancer genomes with high-throughput linked-read sequencing
- PMID: 26829319
- PMCID: PMC4786454
- DOI: 10.1038/nbt.3432
Haplotyping germline and cancer genomes with high-throughput linked-read sequencing
Abstract
Haplotyping of human chromosomes is a prerequisite for cataloguing the full repertoire of genetic variation. We present a microfluidics-based, linked-read sequencing technology that can phase and haplotype germline and cancer genomes using nanograms of input DNA. This high-throughput platform prepares barcoded libraries for short-read sequencing and computationally reconstructs long-range haplotype and structural variant information. We generate haplotype blocks in a nuclear trio that are concordant with expected inheritance patterns and phase a set of structural variants. We also resolve the structure of the EML4-ALK gene fusion in the NCI-H2228 cancer cell line using phased exome sequencing. Finally, we assign genetic aberrations to specific megabase-scale haplotypes generated from whole-genome sequencing of a primary colorectal adenocarcinoma. This approach resolves haplotype information using up to 100 times less genomic DNA than some methods and enables the accurate detection of structural variants.
Conflict of interest statement
The following authors, as listed by initials, are employees of 10× Genomics: G.X.Y.Z., M.S., M.J., C.M.H., S.K.P, D.A.M., L.M., J.M.T., P.A.M., P.W.W., R.B., A.J.M., Y.L., P.B., A.D.P., A.J.L., P.J.M, G.M.V., P.H., L.M., M.L., L.G., A.W., D.E.B., S.W.S., K.P.B., P.V. P., S.K., G.K.L., D.L.S., J.P.D., I.W., H.S.O, J.Y.L., Z.K.B., K.M.G, W.H.H., G.P.D., Z.W.B., F.M., N.O.K., R.T.W., J.A.B., S.P.G., A.P.K., C.B., A.N.F., A.C., S.S., K.D.N., B.J.H.
Figures



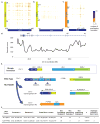
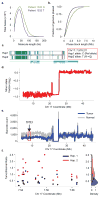
Comment in
-
Haplotypes drop by drop.Nat Biotechnol. 2016 Mar;34(3):296-8. doi: 10.1038/nbt.3500. Nat Biotechnol. 2016. PMID: 26963554 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

