Genome-Wide Analysis of the Lysine Biosynthesis Pathway Network during Maize Seed Development
- PMID: 26829553
- PMCID: PMC4734768
- DOI: 10.1371/journal.pone.0148287
Genome-Wide Analysis of the Lysine Biosynthesis Pathway Network during Maize Seed Development
Abstract
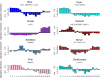
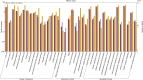
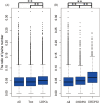
Lysine is one of the most limiting essential amino acids for humans and livestock. The nutritional value of maize (Zea mays L.) is reduced by its poor lysine content. To better understand the lysine biosynthesis pathway in maize seed, we conducted a genome-wide analysis of the genes involved in lysine biosynthesis. We identified lysine biosynthesis pathway genes (LBPGs) and investigated whether a diaminopimelate pathway variant exists in maize. We analyzed two genes encoding the key enzyme dihydrodipicolinate synthase, and determined that they contribute differently to lysine synthesis during maize seed development. A coexpression network of LBPGs was constructed using RNA-sequencing data from 21 developmental stages of B73 maize seed. We found a large set of genes encoding ribosomal proteins, elongation factors and zein proteins that were coexpressed with LBPGs. The coexpressed genes were enriched in cellular metabolism terms and protein related terms. A phylogenetic analysis of the LBPGs from different plant species revealed different relationships. Additionally, six transcription factor (TF) families containing 13 TFs were identified as the Hub TFs of the LBPGs modules. Several expression quantitative trait loci of LBPGs were also identified. Our results should help to elucidate the lysine biosynthesis pathway network in maize seed.
Conflict of interest statement
Figures



Similar articles
-
The dynamic transcriptome of waxy maize (Zea mays L. sinensis Kulesh) during seed development.Genes Genomics. 2020 Sep;42(9):997-1010. doi: 10.1007/s13258-020-00967-z. Epub 2020 Jul 16. Genes Genomics. 2020. PMID: 32676852
-
Seed-specific expression of a lysine-rich protein gene, GhLRP, from cotton significantly increases the lysine content in maize seeds.Int J Mol Sci. 2014 Mar 27;15(4):5350-65. doi: 10.3390/ijms15045350. Int J Mol Sci. 2014. PMID: 24681583 Free PMC article.
-
Distinct tissue-specific transcriptional regulation revealed by gene regulatory networks in maize.BMC Plant Biol. 2018 Jun 7;18(1):111. doi: 10.1186/s12870-018-1329-y. BMC Plant Biol. 2018. PMID: 29879919 Free PMC article.
-
Seed-specific expression of the lysine-rich protein gene sb401 significantly increases both lysine and total protein content in maize seeds.Food Nutr Bull. 2005 Dec;26(4):427-31. Food Nutr Bull. 2005. PMID: 16465991 Review.
-
The regulation of zein biosynthesis in maize endosperm.Theor Appl Genet. 2020 May;133(5):1443-1453. doi: 10.1007/s00122-019-03520-z. Epub 2020 Jan 2. Theor Appl Genet. 2020. PMID: 31897513 Review.
Cited by
-
Characterization of recombinant dihydrodipicolinate synthase from the bread wheat Triticum aestivum.Planta. 2018 Aug;248(2):381-391. doi: 10.1007/s00425-018-2894-x. Epub 2018 May 9. Planta. 2018. PMID: 29744651
-
Recent advances in exploring transcriptional regulatory landscape of crops.Front Plant Sci. 2024 Jun 5;15:1421503. doi: 10.3389/fpls.2024.1421503. eCollection 2024. Front Plant Sci. 2024. PMID: 38903438 Free PMC article. Review.
-
GWAS and WGCNA uncover hub genes controlling salt tolerance in maize (Zea mays L.) seedlings.Theor Appl Genet. 2021 Oct;134(10):3305-3318. doi: 10.1007/s00122-021-03897-w. Epub 2021 Jul 4. Theor Appl Genet. 2021. PMID: 34218289
-
Dissecting the genetic architecture of polygenic nutritional traits in maize through meta-QTL analysis.Food Chem (Oxf). 2025 Apr 24;10:100256. doi: 10.1016/j.fochms.2025.100256. eCollection 2025 Jun. Food Chem (Oxf). 2025. PMID: 40336954 Free PMC article.
-
Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor.Sci Rep. 2018 May 21;8(1):7936. doi: 10.1038/s41598-018-26291-x. Sci Rep. 2018. PMID: 29786696 Free PMC article.
References
-
- Bright SW, Shewry PR, Kasarda DD. Improvement of protein quality in cereals. Crit Rev Plant Sci. 1983;1:49–93.
-
- Prasanna BM, Vasal SK, Kassahun B, Singh NN. Quality protein maize. Current Science Association. 2001;81:1308–1319.
-
- Xu H, Andi B, Qian J, West AH, Cook PF. The α-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem Biophys. 2006;46:43–64. - PubMed
-
- Velasco AM, Leguina JI, Lazcano A. Molecular evolution of the lysine biosynthetic pathways. J Mol Evol. 2002;55:445–459. - PubMed
-
- Scapin G, Blanchard JS. Enzymology of bacterial lysine biosynthesis. Adv Enzymol Relat Areas Mol Biol. 1998;72:279–324. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

