IFNs Modify the Proteome of Legionella-Containing Vacuoles and Restrict Infection Via IRG1-Derived Itaconic Acid
- PMID: 26829557
- PMCID: PMC4734697
- DOI: 10.1371/journal.ppat.1005408
IFNs Modify the Proteome of Legionella-Containing Vacuoles and Restrict Infection Via IRG1-Derived Itaconic Acid
Abstract
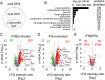
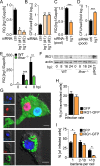
Macrophages can be niches for bacterial pathogens or antibacterial effector cells depending on the pathogen and signals from the immune system. Here we show that type I and II IFNs are master regulators of gene expression during Legionella pneumophila infection, and activators of an alveolar macrophage-intrinsic immune response that restricts bacterial growth during pneumonia. Quantitative mass spectrometry revealed that both IFNs substantially modify Legionella-containing vacuoles, and comparative analyses reveal distinct subsets of transcriptionally and spatially IFN-regulated proteins. Immune-responsive gene (IRG)1 is induced by IFNs in mitochondria that closely associate with Legionella-containing vacuoles, and mediates production of itaconic acid. This metabolite is bactericidal against intravacuolar L. pneumophila as well as extracellular multidrug-resistant Gram-positive and -negative bacteria. Our study explores the overall role IFNs play in inducing substantial remodeling of bacterial vacuoles and in stimulating production of IRG1-derived itaconic acid which targets intravacuolar pathogens. IRG1 or its product itaconic acid might be therapeutically targetable to fight intracellular and drug-resistant bacteria.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

