Development of an Analysis Pipeline Characterizing Multiple Hypervariable Regions of 16S rRNA Using Mock Samples
- PMID: 26829716
- PMCID: PMC4734828
- DOI: 10.1371/journal.pone.0148047
Development of an Analysis Pipeline Characterizing Multiple Hypervariable Regions of 16S rRNA Using Mock Samples
Abstract
Objectives: There is much speculation on which hypervariable region provides the highest bacterial specificity in 16S rRNA sequencing. The optimum solution to prevent bias and to obtain a comprehensive view of complex bacterial communities would be to sequence the entire 16S rRNA gene; however, this is not possible with second generation standard library design and short-read next-generation sequencing technology.
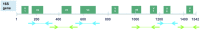
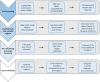
Methods: This paper examines a new process using seven hypervariable or V regions of the 16S rRNA (six amplicons: V2, V3, V4, V6-7, V8, and V9) processed simultaneously on the Ion Torrent Personal Genome Machine (Life Technologies, Grand Island, NY). Four mock samples were amplified using the 16S Ion Metagenomics Kit™ (Life Technologies) and their sequencing data is subjected to a novel analytical pipeline.
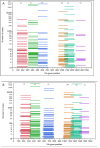
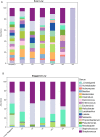
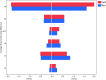
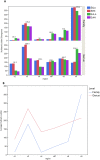
Results: Results are presented at family and genus level. The Kullback-Leibler divergence (DKL), a measure of the departure of the computed from the nominal bacterial distribution in the mock samples, was used to infer which region performed best at the family and genus levels. Three different hypervariable regions, V2, V4, and V6-7, produced the lowest divergence compared to the known mock sample. The V9 region gave the highest (worst) average DKL while the V4 gave the lowest (best) average DKL. In addition to having a high DKL, the V9 region in both the forward and reverse directions performed the worst finding only 17% and 53% of the known family level and 12% and 47% of the genus level bacteria, while results from the forward and reverse V4 region identified all 17 family level bacteria.
Conclusions: The results of our analysis have shown that our sequencing methods using 6 hypervariable regions of the 16S rRNA and subsequent analysis is valid. This method also allowed for the assessment of how well each of the variable regions might perform simultaneously. Our findings will provide the basis for future work intended to assess microbial abundance at different time points throughout a clinical protocol.
Conflict of interest statement
Figures

References
-
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94(3):441–448. - PubMed
-
- Salipante SJ, Kawashima T, Rosenthal C, Hoogestraat DR, Cummings LA. Sengupta DJ,et al. Performance comparison of Illumina and ion torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl Environ Microbiol. 2014;80(24):7583–7591. 10.1128/AEM.02206-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

