MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism
- PMID: 26829751
- PMCID: PMC4767685
- DOI: 10.1038/ng.3500
MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism
Abstract
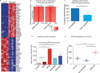
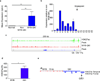
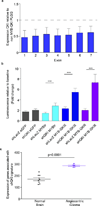
Angiocentric gliomas are pediatric low-grade gliomas (PLGGs) without known recurrent genetic drivers. We performed genomic analysis of new and published data from 249 PLGGs, including 19 angiocentric gliomas. We identified MYB-QKI fusions as a specific and single candidate driver event in angiocentric gliomas. In vitro and in vivo functional studies show that MYB-QKI rearrangements promote tumorigenesis through three mechanisms: MYB activation by truncation, enhancer translocation driving aberrant MYB-QKI expression and hemizygous loss of the tumor suppressor QKI. To our knowledge, this represents the first example of a single driver rearrangement simultaneously transforming cells via three genetic and epigenetic mechanisms in a tumor.
Conflict of interest statement
Conflict of interest statement: The authors declare no conflicts of interest.
Figures





References
References – Main Text
-
- Wang M, et al. Monomorphous angiocentric glioma: a distinctive epileptogenic neoplasm with features of infiltrating astrocytoma ependymoma. J. Neuropathol. Exp. Neurol. 2005;64:875–881. - PubMed
References – Online Methods
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA188228/CA/NCI NIH HHS/United States
- 1R01NS091620/NS/NINDS NIH HHS/United States
- T32 HL007627/HL/NHLBI NIH HHS/United States
- P01 CA142536/CA/NCI NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- P01CA142536/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- K08 NS087118/NS/NINDS NIH HHS/United States
- R01NS085336/NS/NINDS NIH HHS/United States
- R01 NS085336/NS/NINDS NIH HHS/United States
- K08NS087118/NS/NINDS NIH HHS/United States
- F30 CA192725/CA/NCI NIH HHS/United States
- F32 CA180653/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

