Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity
- PMID: 26829754
- PMCID: PMC4768348
- DOI: 10.1038/ng.3501
Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity
Erratum in
-
Corrigendum: Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity.Nat Genet. 2017 Mar 30;49(4):651. doi: 10.1038/ng0417-651b. Nat Genet. 2017. PMID: 28358126 No abstract available.
-
Erratum: Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity.Nat Genet. 2017 May 26;49(6):970. doi: 10.1038/ng0617-970d. Nat Genet. 2017. PMID: 28546578 No abstract available.
Abstract
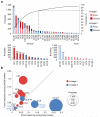
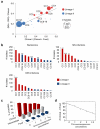
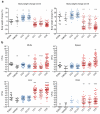
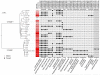
Microbial pathogenesis studies are typically performed with reference strains, thereby overlooking within-species heterogeneity in microbial virulence. Here we integrated human epidemiological and clinical data with bacterial population genomics to harness the biodiversity of the model foodborne pathogen Listeria monocytogenes and decipher the basis of its neural and placental tropisms. Taking advantage of the clonal structure of this bacterial species, we identify clones epidemiologically associated either with food or with human central nervous system (CNS) or maternal-neonatal (MN) listeriosis. The latter clones are also most prevalent in patients without immunosuppressive comorbidities. Strikingly, CNS- and MN-associated clones are hypervirulent in a humanized mouse model of listeriosis. By integrating epidemiological data and comparative genomics, we have uncovered multiple new putative virulence factors and demonstrate experimentally the contribution of the first gene cluster mediating L. monocytogenes neural and placental tropisms. This study illustrates the exceptional power in harnessing microbial biodiversity to identify clinically relevant microbial virulence attributes.
Figures

Comment in
-
Biodiversity and hypervirulence of Listeria monocytogenes.Nat Genet. 2016 Mar;48(3):229-30. doi: 10.1038/ng.3515. Nat Genet. 2016. PMID: 26906682
-
Bacterial GWAS: not just gilding the lily.Nat Rev Microbiol. 2016 Jul;14(7):406. doi: 10.1038/nrmicro.2016.82. Epub 2016 May 31. Nat Rev Microbiol. 2016. PMID: 27241043 No abstract available.
References
-
- Falkow S, Isberg RR, Portnoy DA. The interaction of bacteria with mammalian cells. Annu Rev Cell Biol. 1992;8:333–363. - PubMed
-
- Cossart P, Boquet P, Normark S, Rappuoli R. Cellular microbiology emerging. Science. 1996;271:315–316. - PubMed
-
- Hensel M, et al. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. - PubMed
References for online methods
-
- Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand J Statist. 1979;6:65–70.
-
- Bonferroni CE. Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 1936;8:3–62.
-
- Disson O, et al. Modeling human listeriosis in natural and genetically engineered animals. Nat Protoc. 2009;4:799–810. - PubMed
-
- Criscuolo A, Brisse S. AlienTrimmer: a tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics. 2013;102:500–506. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

