Digital detection of endonuclease mediated gene disruption in the HIV provirus
- PMID: 26829887
- PMCID: PMC4735761
- DOI: 10.1038/srep20064
Digital detection of endonuclease mediated gene disruption in the HIV provirus
Abstract
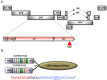
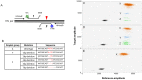
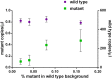
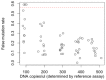
Genome editing by designer nucleases is a rapidly evolving technology utilized in a highly diverse set of research fields. Among all fields, the T7 endonuclease mismatch cleavage assay, or Surveyor assay, is the most commonly used tool to assess genomic editing by designer nucleases. This assay, while relatively easy to perform, provides only a semi-quantitative measure of mutation efficiency that lacks sensitivity and accuracy. We demonstrate a simple droplet digital PCR assay that quickly quantitates a range of indel mutations with detection as low as 0.02% mutant in a wild type background and precision (≤6%CV) and accuracy superior to either mismatch cleavage assay or clonal sequencing when compared to next-generation sequencing. The precision and simplicity of this assay will facilitate comparison of gene editing approaches and their optimization, accelerating progress in this rapidly-moving field.
Figures
Similar articles
-
Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases.G3 (Bethesda). 2015 Jan 7;5(3):407-15. doi: 10.1534/g3.114.015834. G3 (Bethesda). 2015. PMID: 25566793 Free PMC article.
-
Comparison of digital PCR platforms and semi-nested qPCR as a tool to determine the size of the HIV reservoir.Sci Rep. 2015 Sep 9;5:13811. doi: 10.1038/srep13811. Sci Rep. 2015. PMID: 26350506 Free PMC article.
-
[The detection of provirus in lymphocyte DNA. The monitoring of HIV infection at the genetic level].Vopr Virusol. 1994 May-Jun;39(3):107-10. Vopr Virusol. 1994. PMID: 8091747 Russian.
-
Flow cytometry in situ polymerase chain reaction as an innovative approach to HIV-1 proviral DNA detection.Eur J Histochem. 1998;42(2):137-41. Eur J Histochem. 1998. PMID: 9728291 Review.
-
[Progress in digital PCR technology and application].Sheng Wu Gong Cheng Xue Bao. 2017 Feb 25;33(2):170-177. doi: 10.13345/j.cjb.160269. Sheng Wu Gong Cheng Xue Bao. 2017. PMID: 28956373 Review. Chinese.
Cited by
-
Characterization of the HIV-1 transcription profile after romidepsin administration in ART-suppressed individuals.AIDS. 2019 Mar 1;33(3):425-431. doi: 10.1097/QAD.0000000000002083. AIDS. 2019. PMID: 30531314 Free PMC article. Clinical Trial.
-
A One-Step PCR-Based Assay to Evaluate the Efficiency and Precision of Genomic DNA-Editing Tools.Mol Ther Methods Clin Dev. 2017 Mar 8;5:43-50. doi: 10.1016/j.omtm.2017.03.001. eCollection 2017 Jun 16. Mol Ther Methods Clin Dev. 2017. PMID: 28480303 Free PMC article.
-
A simple and efficient method to visualize and quantify the efficiency of chromosomal mutations from genome editing.Sci Rep. 2016 Oct 17;6:35488. doi: 10.1038/srep35488. Sci Rep. 2016. PMID: 27748423 Free PMC article.
-
Pharmacodynamics of anti-HIV gene therapy using viral vectors and targeted endonucleases.J Antimicrob Chemother. 2016 Aug;71(8):2089-99. doi: 10.1093/jac/dkw104. Epub 2016 Apr 18. J Antimicrob Chemother. 2016. PMID: 27090632 Free PMC article.
-
Cell and gene therapy strategies to eradicate HIV reservoirs.Curr Opin HIV AIDS. 2016 Jul;11(4):442-9. doi: 10.1097/COH.0000000000000284. Curr Opin HIV AIDS. 2016. PMID: 27031009 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

