Touch and go: nuclear proteolysis in the regulation of metabolic genes and cancer
- PMID: 26832397
- PMCID: PMC4833644
- DOI: 10.1002/1873-3468.12087
Touch and go: nuclear proteolysis in the regulation of metabolic genes and cancer
Abstract
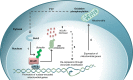
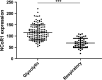
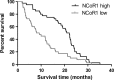
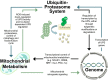
The recruitment of transcription factors to promoters and enhancers is a critical step in gene regulation. Many of these proteins are quickly removed from DNA after they completed their function. Metabolic genes in particular are dynamically regulated and continuously adjusted to cellular requirements. Transcription factors controlling metabolism are therefore under constant surveillance by the ubiquitin-proteasome system, which can degrade DNA-bound proteins in a site-specific manner. Several of these metabolic transcription factors are critical to cancer cells, as they promote uncontrolled growth and proliferation. This review highlights recent findings in the emerging field of nuclear proteolysis and outlines novel paradigms for cancer treatment, with an emphasis on multiple myeloma.
Keywords: genome regulation; mitochondria; multiple myeloma; nuclear proteostasis; nuclear receptor corepressor 1; ubiquitin-proteasome system.
© The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures

References
-
- Kroemer G and Pouyssegur J (2008) Tumor cell metabolism: cancer's Achilles’ heel. Cancer Cell 13, 472–482. - PubMed
-
- Batra S, Adekola KU, Rosen ST and Shanmugam M (2013) Cancer metabolism as a therapeutic target. Oncology (Williston Park) 27, 460–467. - PubMed
-
- Komander D, Clague MJ and Urbe S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10, 550–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

