Nac1 Coordinates a Sub-network of Pluripotency Factors to Regulate Embryonic Stem Cell Differentiation
- PMID: 26832399
- PMCID: PMC4749452
- DOI: 10.1016/j.celrep.2015.12.101
Nac1 Coordinates a Sub-network of Pluripotency Factors to Regulate Embryonic Stem Cell Differentiation
Abstract
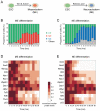
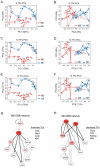
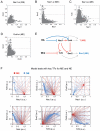
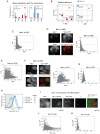
Pluripotent cells give rise to distinct cell types during development and are regulated by often self-reinforcing molecular networks. How such networks allow cells to differentiate is less well understood. Here, we use integrative methods to show that external signals induce reorganization of the mouse embryonic stem cell pluripotency network and that a sub-network of four factors, Nac1, Oct4, Tcf3, and Sox2, regulates their differentiation into the alternative mesendodermal and neuroectodermal fates. In the mesendodermal fate, Nac1 and Oct4 were constrained within quantitative windows, whereas Sox2 and Tcf3 were repressed. In contrast, in the neuroectodermal fate, Sox2 and Tcf3 were constrained while Nac1 and Oct4 were repressed. In addition, we show that Nac1 coordinates differentiation by activating Oct4 and inhibiting both Sox2 and Tcf3. Reorganization of progenitor cell networks around shared factors might be a common differentiation strategy and our integrative approach provides a general methodology for delineating such networks.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Molecular and cellular endocrinology. 2002;195:27–38. - PubMed
-
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

